Precision Metagenomics: Rapid Metagenomic Analyses for Infectious Disease Diagnostics and Public Health Surveillance
- PMID: 28337072
- PMCID: PMC5360386
- DOI: 10.7171/jbt.17-2801-007
Precision Metagenomics: Rapid Metagenomic Analyses for Infectious Disease Diagnostics and Public Health Surveillance
Erratum in
-
[No title available]J Biomol Tech. 2017 Jul;28(2):95. doi: 10.7171/jbt.17-2801-007CX. Epub 2017 Jun 8. J Biomol Tech. 2017. PMID: 28630599 Free PMC article.
Abstract
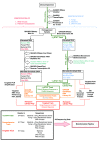
Next-generation sequencing (NGS) technologies have ushered in the era of precision medicine, transforming the way we treat cancer patients and diagnose disease. Concomitantly, the advent of these technologies has created a surge of microbiome and metagenomic studies over the last decade, many of which are focused on investigating the host-gene-microbial interactions responsible for the development and spread of infectious diseases, as well as delineating their key role in maintaining health. As we continue to discover more information about the etiology of infectious diseases, the translational potential of metagenomic NGS methods for treatment and rapid diagnosis is becoming abundantly clear. Here, we present a robust protocol for the implementation and application of "precision metagenomics" across various sequencing platforms for clinical samples. Such a pipeline integrates DNA/RNA extraction, library preparation, sequencing, and bioinformatics analyses for taxonomic classification, antimicrobial resistance (AMR) marker screening, and functional analysis (biochemical and metabolic pathway abundance). Moreover, the pipeline has 3 tracks: STAT for results within 24 h; Comprehensive that affords a more in-depth analysis and takes between 5 and 7 d, but offers antimicrobial resistance information; and Targeted, which also requires 5-7 d, but with more sensitive analysis for specific pathogens. Finally, we discuss the challenges that need to be addressed before full integration in the clinical setting.
Keywords: antimicrobial resistance; microbiome; next-generation sequencing; pathogen detection.
Figures
References
-
- Fishman JA. Infections in immunocompromised hosts and organ transplant recipients: essentials. Liver Transpl 2011;17 (Suppl 3):S34–S37. - PubMed
-
- Vartoukian SR, Palmer RM, Wade WG. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol Lett 2010;309:1–7. - PubMed
-
- Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016;16:161–168. - PubMed
-
- Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006;34:1589–1596. - PubMed
-
- Kaplan RS, Porter ME. How to solve the cost crisis in health care. Harv Bus Rev 2011;89:46–52, 54, 56–61 passim. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
