Dissecting miRNA gene repression on single cell level with an advanced fluorescent reporter system
- PMID: 28338079
- PMCID: PMC5364550
- DOI: 10.1038/srep45197
Dissecting miRNA gene repression on single cell level with an advanced fluorescent reporter system
Abstract
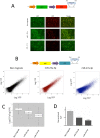
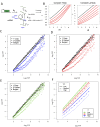
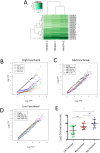
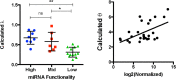
Despite major advances on miRNA profiling and target predictions, functional readouts for endogenous miRNAs are limited and frequently lead to contradicting conclusions. Numerous approaches including functional high-throughput and miRISC complex evaluations suggest that the functional miRNAome differs from the predictions based on quantitative sRNA profiling. To resolve the apparent contradiction of expression versus function, we generated and applied a fluorescence reporter gene assay enabling single cell analysis. This approach integrates and adapts a mathematical model for miRNA-driven gene repression. This model predicts three distinct miRNA-groups with unique repression activities (low, mid and high) governed not just by expression levels but also by miRNA/target-binding capability. Here, we demonstrate the feasibility of the system by applying controlled concentrations of synthetic siRNAs and in parallel, altering target-binding capability on corresponding reporter-constructs. Furthermore, we compared miRNA-profiles with the modeled predictions of 29 individual candidates. We demonstrate that expression levels only partially reflect the miRNA function, fitting to the model-projected groups of different activities. Furthermore, we demonstrate that subcellular localization of miRNAs impacts functionality. Our results imply that miRNA profiling alone cannot define their repression activity. The gene regulatory function is a dynamic and complex process beyond a minimalistic conception of "highly expressed equals high repression".
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

