Halo-free Phase Contrast Microscopy
- PMID: 28338086
- PMCID: PMC5364506
- DOI: 10.1038/srep44034
Halo-free Phase Contrast Microscopy
Abstract
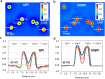
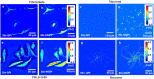
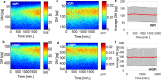
We present a new approach for retrieving halo-free phase contrast microscopy (hfPC) images by upgrading the conventional PC microscope with an external interferometric module, which generates sufficient data for reversing the halo artifact. Acquiring four independent intensity images, our approach first measures haloed phase maps of the sample. We solve for the halo-free sample transmission function by using a physical model of the image formation under partial spatial coherence. Using this halo-free sample transmission, we can numerically generate artifact-free PC images. Furthermore, this transmission can be further used to obtain quantitative information about the sample, e.g., the thickness with known refractive indices, dry mass of live cells during their cycles. We tested our hfPC method on various control samples, e.g., beads, pillars and validated its potential for biological investigation by imaging live HeLa cells, red blood cells, and neurons.
Conflict of interest statement
G.P. has financial interest in Phi Optics, Inc., a company developing quantitative phase imaging technology for materials and life science applications.
Figures




References
-
- Evanko D., Heinrichs A. & Rosenthal C. Milestones in light microscopy. Nature Cell Biol. 11, S5–S20 (2009). - PubMed
-
- Abbe E. Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung. Archiv für mikroskopische Anatomie 9, 413–418 (1873).
-
- Hell S. W. & Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Optics letters 19, 780–782 (1994). - PubMed
-
- Pluta M. Advanced light microscopy. (Polish Scientific Publishers, 1988).
-
- Popescu G. Quantitative phase imaging of cells and tissues. (McGraw-Hill, 2011).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

