Inhibitors of Protein Methyltransferases and Demethylases
- PMID: 28338320
- PMCID: PMC5610952
- DOI: 10.1021/acs.chemrev.6b00801
Inhibitors of Protein Methyltransferases and Demethylases
Abstract
Post-translational modifications of histones by protein methyltransferases (PMTs) and histone demethylases (KDMs) play an important role in the regulation of gene expression and transcription and are implicated in cancer and many other diseases. Many of these enzymes also target various nonhistone proteins impacting numerous crucial biological pathways. Given their key biological functions and implications in human diseases, there has been a growing interest in assessing these enzymes as potential therapeutic targets. Consequently, discovering and developing inhibitors of these enzymes has become a very active and fast-growing research area over the past decade. In this review, we cover the discovery, characterization, and biological application of inhibitors of PMTs and KDMs with emphasis on key advancements in the field. We also discuss challenges, opportunities, and future directions in this emerging, exciting research field.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




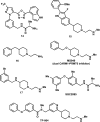
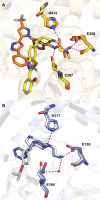
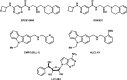
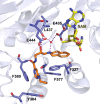

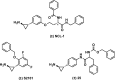
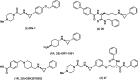
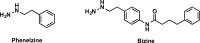
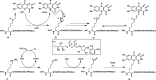
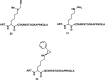
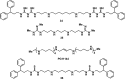
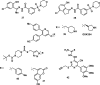
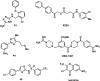
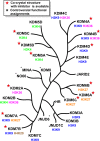

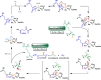

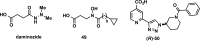
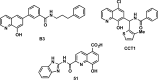

References
-
- Allis C. D.; Jenuwein T.; Reinberg D.. Epigenetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, N.Y., 2007; p. 502.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

