Improving a full-text search engine: the importance of negation detection and family history context to identify cases in a biomedical data warehouse
- PMID: 28339516
- PMCID: PMC7651926
- DOI: 10.1093/jamia/ocw144
Improving a full-text search engine: the importance of negation detection and family history context to identify cases in a biomedical data warehouse
Abstract
Objective: The repurposing of electronic health records (EHRs) can improve clinical and genetic research for rare diseases. However, significant information in rare disease EHRs is embedded in the narrative reports, which contain many negated clinical signs and family medical history. This paper presents a method to detect family history and negation in narrative reports and evaluates its impact on selecting populations from a clinical data warehouse (CDW).
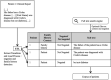
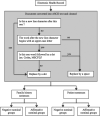
Materials and methods: We developed a pipeline to process 1.6 million reports from multiple sources. This pipeline is part of the load process of the Necker Hospital CDW.
Results: We identified patients with "Lupus and diarrhea," "Crohn's and diabetes," and "NPHP1" from the CDW. The overall precision, recall, specificity, and F-measure were 0.85, 0.98, 0.93, and 0.91, respectively.
Conclusion: The proposed method generates a highly accurate identification of cases from a CDW of rare disease EHRs.
Keywords: data warehouse; electronic health records; natural language processing; rare diseases; search engine.
© The Author 2016. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For Permissions, please email: journals.permissions@oup.com
Figures

References
-
- Cuggia M, Garcelon N, Campillo-Gimenez B et al. Roogle: an information retrieval engine for clinical data warehouse. Stud Health Technol Inform. 2011;169:584–8. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

