Gene and protein analysis reveals that p53 pathway is functionally inactivated in cytogenetically normal Acute Myeloid Leukemia and Acute Promyelocytic Leukemia
- PMID: 28340577
- PMCID: PMC5423421
- DOI: 10.1186/s12920-017-0249-2
Gene and protein analysis reveals that p53 pathway is functionally inactivated in cytogenetically normal Acute Myeloid Leukemia and Acute Promyelocytic Leukemia
Abstract
Background: Mechanisms that inactivate the p53 pathway in Acute Myeloid Leukemia (AML), other than rare mutations, are still not well understood.
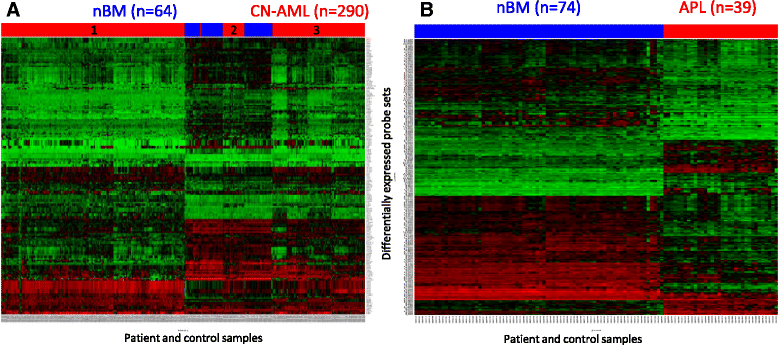
Methods: We performed a bioinformatics study of the p53 pathway function at the gene expression level on our collection of 1153 p53-pathway related genes. Publically available Affymetrix data of 607 de-novo AML patients at diagnosis were analyzed according to the patients cytogenetic, FAB and molecular mutations subtypes. We further investigated the functional status of the p53 pathway in cytogenetically normal AML (CN-AML) and Acute Promyelocytic Leukemia (APL) patients using bioinformatics, Real-Time PCR and immunohistochemistry.
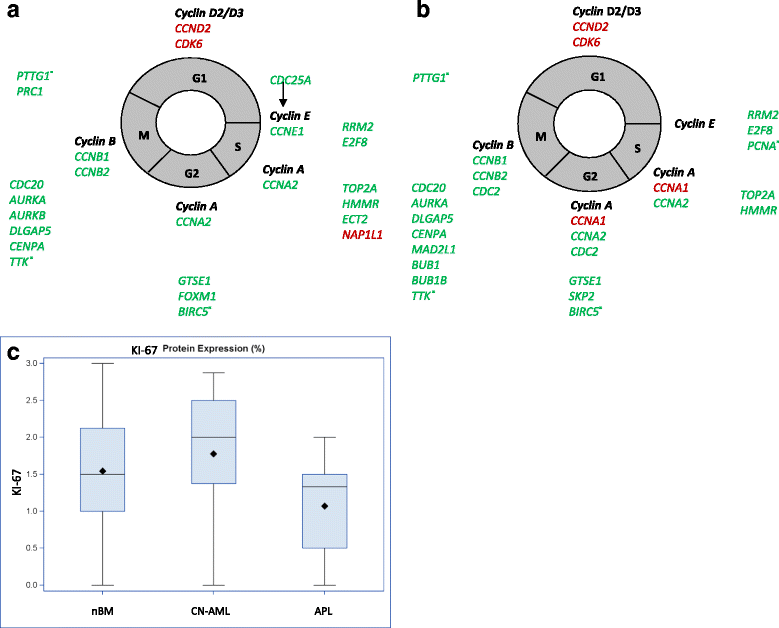
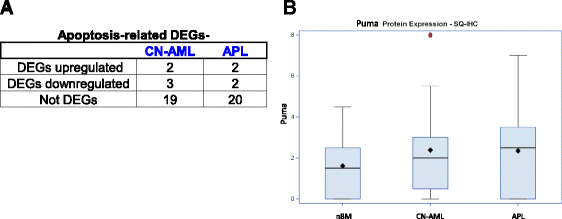
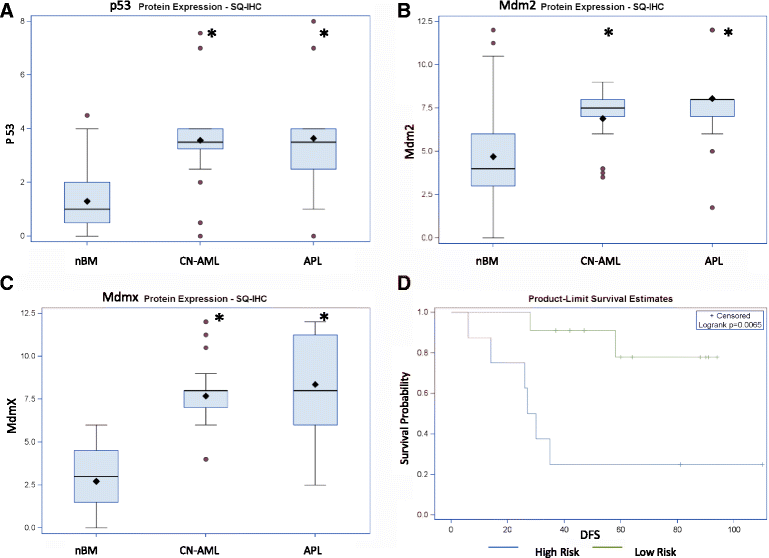
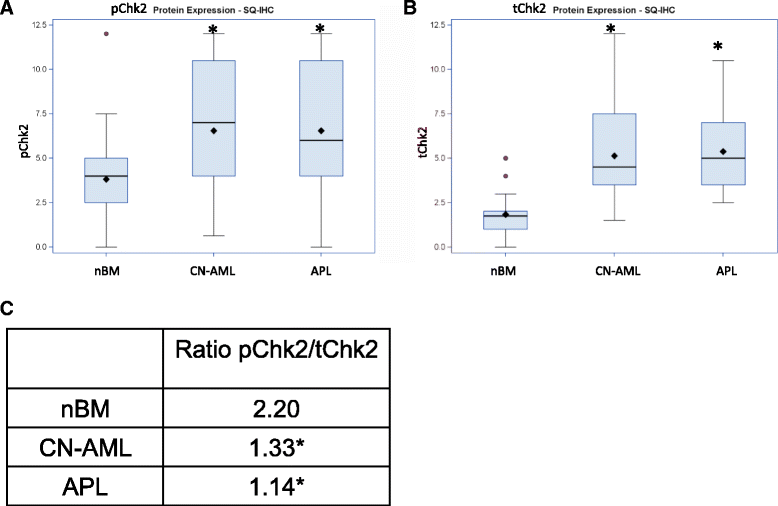
Results: We revealed significant and differential alterations of p53 pathway-related gene expression in most of the AML subtypes. We found that p53 pathway-related gene expression was not correlated with the accepted grouping of AML subtypes such as by cytogenetically-based prognosis, morphological stage or by the type of molecular mutation. Our bioinformatic analysis revealed that p53 is not functional in CN-AML and APL blasts at inducing its most important functional outcomes: cell cycle arrest, apoptosis, DNA repair and oxidative stress defense. We revealed transcriptional downregulation of important p53 acetyltransferases in both CN-AML and APL, accompanied by increased Mdmx protein expression and inadequate Chk2 protein activation.
Conclusions: Our bioinformatic analysis demonstrated that p53 pathway is differentially inactivated in different AML subtypes. Focused gene and protein analysis of p53 pathway in CN-AML and APL patients imply that functional inactivation of p53 protein can be attributed to its impaired acetylation. Our analysis indicates the need in further accurate evaluation of p53 pathway functioning and regulation in distinct subtypes of AML.
Figures
References
-
- Acute Myeloid Leukemia. Leukemia and Lymphoma Society. 2015. www.sllcanada.org/sites/default/files/file_assets/PS32_AML_Booklet_FINAL....
-
- Foran JM. New prognostic markers in acute myeloid leukemia: perspective from the clinic. Hematology Am Soc Hematol Educ Program. 2010;2010:47–55. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

