MeDeCom: discovery and quantification of latent components of heterogeneous methylomes
- PMID: 28340624
- PMCID: PMC5366155
- DOI: 10.1186/s13059-017-1182-6
MeDeCom: discovery and quantification of latent components of heterogeneous methylomes
Abstract
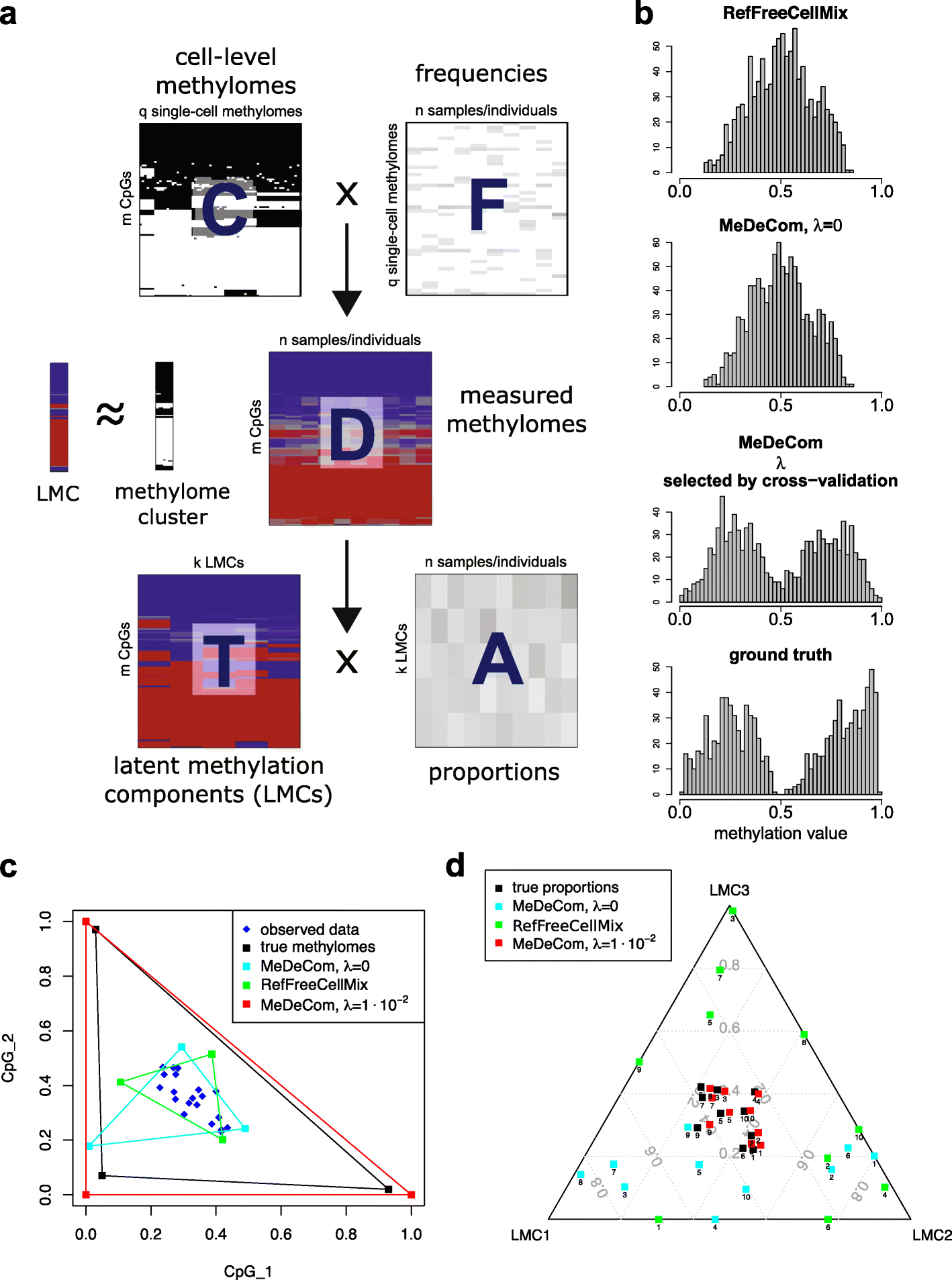
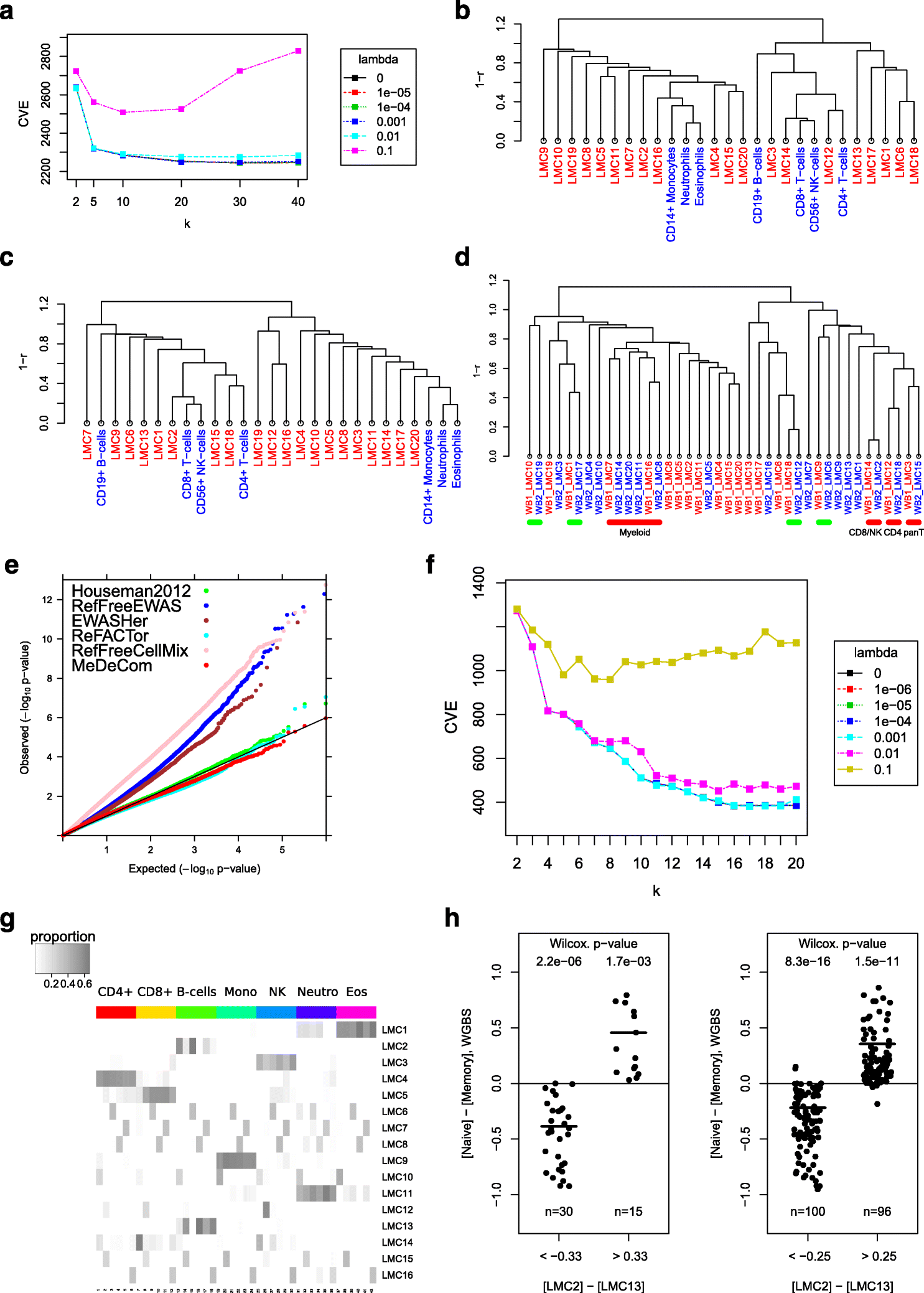
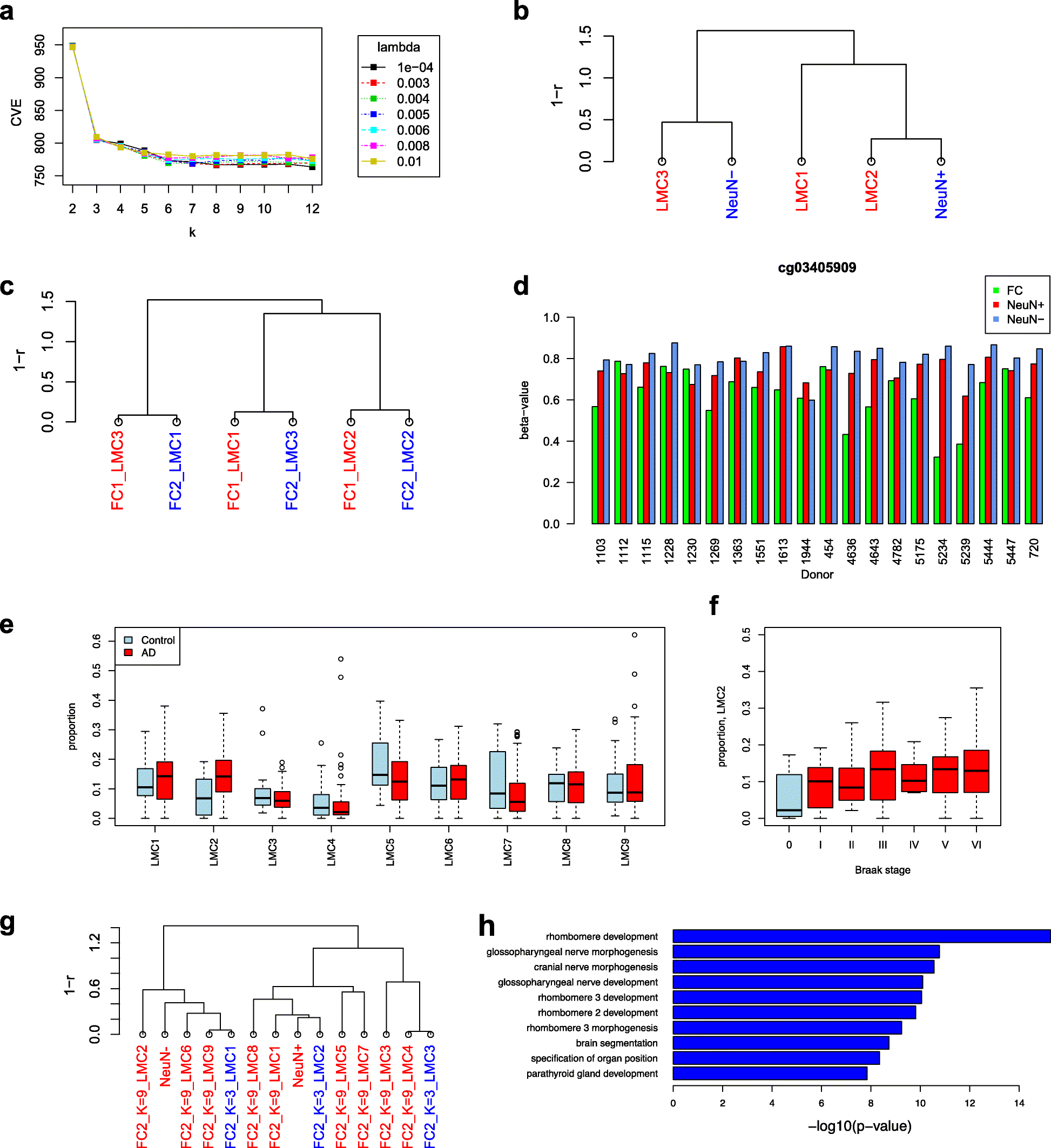
It is important for large-scale epigenomic studies to determine and explore the nature of hidden confounding variation, most importantly cell composition. We developed MeDeCom as a novel reference-free computational framework that allows the decomposition of complex DNA methylomes into latent methylation components and their proportions in each sample. MeDeCom is based on constrained non-negative matrix factorization with a new biologically motivated regularization function. It accurately recovers cell-type-specific latent methylation components and their proportions. MeDeCom is a new unsupervised tool for the exploratory study of the major sources of methylation variation, which should lead to a deeper understanding and better biological interpretation.
Keywords: Cell heterogeneity; DNA methylation; DNA methylome; Deconvolution; Epigenetics; Matrix factorization.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

