Lung Adenocarcinoma and Squamous Cell Carcinoma Gene Expression Subtypes Demonstrate Significant Differences in Tumor Immune Landscape
- PMID: 28341226
- PMCID: PMC6557266
- DOI: 10.1016/j.jtho.2017.03.010
Lung Adenocarcinoma and Squamous Cell Carcinoma Gene Expression Subtypes Demonstrate Significant Differences in Tumor Immune Landscape
Abstract
Introduction: Molecular subtyping of lung adenocarcinoma (AD) and lung squamous cell carcinoma (SCC) reveal biologically diverse tumors that vary in their genomic and clinical attributes.
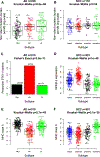
Methods: Published immune cell signatures and several lung AD and SCC gene expression data sets, including The Cancer Genome Atlas, were used to examine immune response in relation to AD and SCC expression subtypes. Expression of immune cell populations and other immune related genes, including CD274 molecule gene (CD274) (programmed death ligand 1), was investigated in the tumor microenvironment relative to the expression subtypes of the AD (terminal respiratory unit, proximal proliferative, and proximal inflammatory) and SCC (primitive, classical, secretory, and basal) subtypes.
Results: Lung AD and SCC expression subtypes demonstrated significant differences in tumor immune landscape. The proximal proliferative subtype of AD demonstrated low immune cell expression among ADs whereas the secretory subtype showed elevated immune cell expression among SCCs. Tumor expression subtype was a better predictor of immune cell expression than CD274 (programmed death ligand 1) in SCC tumors but was a comparable predictor in AD tumors. Nonsilent mutation burden was not correlated with immune cell expression across subtypes; however, major histocompatibility complex class II gene expression was highly correlated with immune cell expression. Increased immune and major histocompatibility complex II gene expression was associated with improved survival in the terminal respiratory unit and proximal inflammatory subtypes of AD and in the primitive subtype of SCC.
Conclusions: Molecular expression subtypes of lung AD and SCC demonstrate key and reproducible differences in immune host response. Evaluation of tumor expression subtypes as potential biomarkers for immunotherapy should be investigated.
Keywords: Adenocarcinoma; Gene expression; Immune response; Lung cancer; PD-L1; Squamous cell carcinoma.
Copyright © 2017 International Association for the Study of Lung Cancer. Published by Elsevier Inc. All rights reserved.
Figures




Comment in
-
Immune Signatures of Non-Small Cell Lung Cancer.J Thorac Oncol. 2017 Jun;12(6):913-915. doi: 10.1016/j.jtho.2017.04.008. J Thorac Oncol. 2017. PMID: 28532560 No abstract available.
References
-
- Hayes DN, Monti S, Parmigiani G, et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol. 2006;24:5079–5090. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

