Comprehensive Determination of Prostate Tumor ETS Gene Status in Clinical Samples Using the CLIA Decipher Assay
- PMID: 28341589
- PMCID: PMC5417038
- DOI: 10.1016/j.jmoldx.2017.01.007
Comprehensive Determination of Prostate Tumor ETS Gene Status in Clinical Samples Using the CLIA Decipher Assay
Abstract
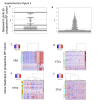
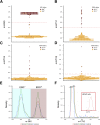
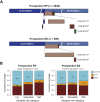
ETS family gene fusions are common in prostate cancer and molecularly define a tumor subset. ERG is the most commonly rearranged, leading to its overexpression, followed by ETV1, ETV4, and ETV5, and these alterations are generally mutually exclusive. We validated the Decipher prostate cancer assay to detect ETS alterations in a Clinical Laboratory Improvement Amendments-accredited laboratory. Benchmarking against ERG immunohistochemistry and ETV1/4/5 RNA in situ hybridization, we examined the accuracy, precision, and reproducibility of gene expression ETS models using formalin-fixed, paraffin-embedded samples. The m-ERG model achieved an area under curve of 95%, with 93% sensitivity and 98% specificity to predict ERG immunohistochemistry status. The m-ETV1, -ETV4, and -ETV5 models achieved areas under curve of 98%, 88%, and 99%, respectively. The models had 100% robustness for ETS status, and scores were highly correlated across sample replicates. Models predicted 41.5% of a prospective radical prostatectomy cohort (n = 4036) to be ERG+, 6.3% ETV1+, 1% ETV4+, and 0.4% ETV5+. Of prostate tumor biopsy samples (n = 509), 41.2% were ERG+, 8.6% ETV1+, 0.4% ETV4+, and none ETV5+. Higher Decipher risk status tumors were more likely to be ETS+ (ERG or ETV1/4/5) in the radical prostatectomy and the biopsy cohorts (P < 0.05). These results support the utility of microarray-based ETS status prediction models for molecular classification of prostate tumors.
Copyright © 2017 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J.E., Shah R.B., Pienta K.J., Rubin M.A., Chinnaiyan A.M. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. - PubMed
-
- Tomlins S.A., Mehra R., Rhodes D.R., Smith L.R., Roulston D., Helgeson B.E., Cao X., Wei J.T., Rubin M.A., Shah R.B., Chinnaiyan A.M. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. - PubMed
-
- Tomlins S.A., Laxman B., Dhanasekaran S.M., Helgeson B.E., Cao X., Morris D.S., Menon A., Jing X., Cao Q., Han B., Yu J., Wang L., Montie J.E., Rubin M.A., Pienta K.J., Roulston D., Shah R.B., Varambally S., Mehra R., Chinnaiyan A.M. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. - PubMed
-
- Paulo P., Barros-Silva J.D., Ribeiro F.R., Ramalho-Carvalho J., Jeronimo C., Henrique R., Lind G.E., Skotheim R.I., Lothe R.A., Teixeira M.R. FLI1 is a novel ETS transcription factor involved in gene fusions in prostate cancer. Genes Chromosomes Cancer. 2012;51:240–249. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

