Estimating Seven Coefficients of Pairwise Relatedness Using Population-Genomic Data
- PMID: 28341647
- PMCID: PMC5419463
- DOI: 10.1534/genetics.116.190660
Estimating Seven Coefficients of Pairwise Relatedness Using Population-Genomic Data
Abstract
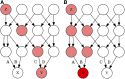
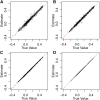
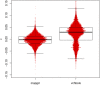
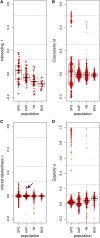
Population structure can be described by genotypic-correlation coefficients between groups of individuals, the most basic of which are the pairwise relatedness coefficients between any two individuals. There are nine pairwise relatedness coefficients in the most general model, and we show that these can be reduced to seven coefficients for biallelic loci. Although all nine coefficients can be estimated from pedigrees, six coefficients have been beyond empirical reach. We provide a numerical optimization procedure that estimates all seven reduced coefficients from population-genomic data. Simulations show that the procedure is nearly unbiased, even at 3× coverage, and errors in five of the seven coefficients are statistically uncorrelated. The remaining two coefficients have a negative correlation of errors, but their sum provides an unbiased assessment of the overall correlation of heterozygosity between two individuals. Application of these new methods to four populations of the freshwater crustacean Daphnia pulex reveal the occurrence of half siblings in our samples, as well as a number of identical individuals that are likely obligately asexual clone mates. Statistically significant negative estimates of these pairwise relatedness coefficients, including inbreeding coefficients that were typically negative, underscore the difficulties that arise when interpreting genotypic correlations as estimations of the probability that alleles are identical by descent.
Keywords: coancestry; identity by descent; population genomics; population structure; relatedness.
Copyright © 2017 by the Genetics Society of America.
Figures
References
-
- Cockerham C., 1983. Covariances of relatives from self-fertilization. Crop Sci. 23: 1177–1180.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

