Accuracy of Demographic Inferences from the Site Frequency Spectrum: The Case of the Yoruba Population
- PMID: 28341655
- PMCID: PMC5419487
- DOI: 10.1534/genetics.116.192708
Accuracy of Demographic Inferences from the Site Frequency Spectrum: The Case of the Yoruba Population
Abstract
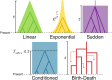
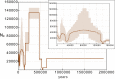
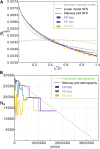
Some methods for demographic inference based on the observed genetic diversity of current populations rely on the use of summary statistics such as the Site Frequency Spectrum (SFS). Demographic models can be either model-constrained with numerous parameters, such as growth rates, timing of demographic events, and migration rates, or model-flexible, with an unbounded collection of piecewise constant sizes. It is still debated whether demographic histories can be accurately inferred based on the SFS. Here, we illustrate this theoretical issue on an example of demographic inference for an African population. The SFS of the Yoruba population (data from the 1000 Genomes Project) is fit to a simple model of population growth described with a single parameter (e.g., founding time). We infer a time to the most recent common ancestor of 1.7 million years (MY) for this population. However, we show that the Yoruba SFS is not informative enough to discriminate between several different models of growth. We also show that for such simple demographies, the fit of one-parameter models outperforms the stairway plot, a recently developed model-flexible method. The use of this method on simulated data suggests that it is biased by the noise intrinsically present in the data.
Keywords: coalescent theory; human demography; model identifiability; site frequency spectrum.
Copyright © 2017 by the Genetics Society of America.
Figures


References
-
- Atkinson Q. D., Gray R. D., Drummond A. J., 2008. mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Mol. Biol. Evol. 25(2): 468–474. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

