asymptoticMK: A Web-Based Tool for the Asymptotic McDonald-Kreitman Test
- PMID: 28341700
- PMCID: PMC5427504
- DOI: 10.1534/g3.117.039693
asymptoticMK: A Web-Based Tool for the Asymptotic McDonald-Kreitman Test
Abstract
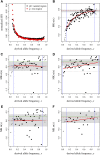
The McDonald-Kreitman (MK) test is a widely used method for quantifying the role of positive selection in molecular evolution. One key shortcoming of this test lies in its sensitivity to the presence of slightly deleterious mutations, which can severely bias its estimates. An asymptotic version of the MK test was recently introduced that addresses this problem by evaluating polymorphism levels for different mutation frequencies separately, and then extrapolating a function fitted to that data. Here, we present asymptoticMK, a web-based implementation of this asymptotic MK test. Our web service provides a simple R-based interface into which the user can upload the required data (polymorphism and divergence data for the genomic test region and a neutrally evolving reference region). The web service then analyzes the data and provides plots of the test results. This service is free to use, open-source, and available at http://benhaller.com/messerlab/asymptoticMK.html We provide results from simulations to illustrate the performance and robustness of the asymptoticMK test under a wide range of model parameters.
Keywords: molecular evolution; positive selection; web service.
Copyright © 2017 Haller and Messer.
Figures
Similar articles
-
iMKT: the integrative McDonald and Kreitman test.Nucleic Acids Res. 2019 Jul 2;47(W1):W283-W288. doi: 10.1093/nar/gkz372. Nucleic Acids Res. 2019. PMID: 31081014 Free PMC article.
-
impMKT: the imputed McDonald and Kreitman test, a straightforward correction that significantly increases the evidence of positive selection of the McDonald and Kreitman test at the gene level.G3 (Bethesda). 2022 Sep 30;12(10):jkac206. doi: 10.1093/g3journal/jkac206. G3 (Bethesda). 2022. PMID: 35976111 Free PMC article. Review.
-
Frequent adaptation and the McDonald-Kreitman test.Proc Natl Acad Sci U S A. 2013 May 21;110(21):8615-20. doi: 10.1073/pnas.1220835110. Epub 2013 May 6. Proc Natl Acad Sci U S A. 2013. PMID: 23650353 Free PMC article.
-
The influence of demography and weak selection on the McDonald-Kreitman test: an empirical study in Drosophila.Mol Biol Evol. 2009 Mar;26(3):691-8. doi: 10.1093/molbev/msn297. Epub 2009 Jan 6. Mol Biol Evol. 2009. PMID: 19126864
-
Role of selection in fixation of gene duplications.J Theor Biol. 2006 Mar 21;239(2):141-51. doi: 10.1016/j.jtbi.2005.08.033. Epub 2005 Oct 20. J Theor Biol. 2006. PMID: 16242725 Review.
Cited by
-
Dissecting Genomic Determinants of Positive Selection with an Evolution-Guided Regression Model.Mol Biol Evol. 2022 Jan 7;39(1):msab291. doi: 10.1093/molbev/msab291. Mol Biol Evol. 2022. PMID: 34597406 Free PMC article.
-
Demographic Processes Linked to Genetic Diversity and Positive Selection across a Species' Range.Plant Commun. 2020 Sep 11;1(6):100111. doi: 10.1016/j.xplc.2020.100111. eCollection 2020 Nov 9. Plant Commun. 2020. PMID: 33367266 Free PMC article.
-
Exploiting selection at linked sites to infer the rate and strength of adaptation.Nat Ecol Evol. 2019 Jun;3(6):977-984. doi: 10.1038/s41559-019-0890-6. Epub 2019 May 6. Nat Ecol Evol. 2019. PMID: 31061475 Free PMC article.
-
iMKT: the integrative McDonald and Kreitman test.Nucleic Acids Res. 2019 Jul 2;47(W1):W283-W288. doi: 10.1093/nar/gkz372. Nucleic Acids Res. 2019. PMID: 31081014 Free PMC article.
-
Tree-sequence recording in SLiM opens new horizons for forward-time simulation of whole genomes.Mol Ecol Resour. 2019 Mar;19(2):552-566. doi: 10.1111/1755-0998.12968. Epub 2019 Feb 21. Mol Ecol Resour. 2019. PMID: 30565882 Free PMC article.
References
-
- Andolfatto P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437: 1149–1152. - PubMed
-
- Bustamante C. D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M. T., et al. , 2005. Natural selection on protein-coding genes in the human genome. Nature 437: 1153–1157. - PubMed
-
- Charlesworth J., Eyre-Walker A., 2008. The McDonald-Kreitman test and slightly deleterious mutations. Mol. Biol. Evol. 25: 1007–1015. - PubMed
-
- Clark A. G., Eisen M. B., Smith D. R., Bergman C. M., Oliver B., et al. , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
