Structural landscape of base pairs containing post-transcriptional modifications in RNA
- PMID: 28341704
- PMCID: PMC5435857
- DOI: 10.1261/rna.060749.117
Structural landscape of base pairs containing post-transcriptional modifications in RNA
Abstract
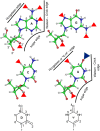
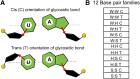
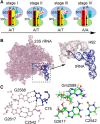
Base pairs involving post-transcriptionally modified nucleobases are believed to play important roles in a wide variety of functional RNAs. Here we present our attempts toward understanding the structural and functional role of naturally occurring modified base pairs using a combination of X-ray crystal structure database analysis, sequence analysis, and advanced quantum chemical methods. Our bioinformatics analysis reveals that despite their presence in all major secondary structural elements, modified base pairs are most prevalent in tRNA crystal structures and most commonly involve guanine or uridine modifications. Further, analysis of tRNA sequences reveals additional examples of modified base pairs at structurally conserved tRNA regions and highlights the conservation patterns of these base pairs in three domains of life. Comparison of structures and binding energies of modified base pairs with their unmodified counterparts, using quantum chemical methods, allowed us to classify the base modifications in terms of the nature of their electronic structure effects on base-pairing. Analysis of specific structural contexts of modified base pairs in RNA crystal structures revealed several interesting scenarios, including those at the tRNA:rRNA interface, antibiotic-binding sites on the ribosome, and the three-way junctions within tRNA. These scenarios, when analyzed in the context of available experimental data, allowed us to correlate the occurrence and strength of modified base pairs with their specific functional roles. Overall, our study highlights the structural importance of modified base pairs in RNA and points toward the need for greater appreciation of the role of modified bases and their interactions, in the context of many biological processes involving RNA.
Keywords: X-ray crystal structures; base-pair parameters; interaction energies; modified base pairs; post-transcriptional modifications.
© 2017 Seelam et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





References
-
- Ahlrichs R, Elliott SD, Huniar U. 1998. Quantum chemistry: large molecules–small computers. Ber Bunsen-Ges Phys Chem 102: 795–804.
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. - PubMed
-
- Bansal M, Bhattacharyya D, Ravi B. 1995. NUPARM and NUCGEN: software for analysis and generation of sequence dependent nucleic acid structures. Comput Appl Biosci 11: 281–287. - PubMed
-
- Becke AD. 1993. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98: 5648–5652.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
