Isolation and characterization of a novel putative human polyomavirus
- PMID: 28342387
- PMCID: PMC9265179
- DOI: 10.1016/j.virol.2017.03.007
Isolation and characterization of a novel putative human polyomavirus
Abstract
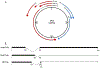
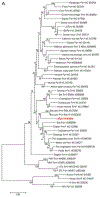
The small double-stranded DNA polyomaviruses (PyVs) form a family of 73 species, whose natural hosts are primarily mammals and birds. So far, 13 PyVs have been isolated in humans, and some of them have clearly been associated with several diseases, including cancer. In this study, we describe the isolation of a novel PyV in human skin using a sensitive degenerate PCR protocol combined with next-generation sequencing. The new virus, named Lyon IARC PyV (LIPyV), has a circular genome of 5269 nucleotides. Phylogenetic analyses showed that LIPyV is related to the raccoon PyV identified in neuroglial tumours in free-ranging raccoons. Analysis of human specimens from cancer-free individuals showed that 9 skin swabs (9/445; 2.0%), 3 oral gargles (3/140; 2.1%), and one eyebrow hair sample (1/439; 0.2%) tested positive for LIPyV. Future biological and epidemiological studies are needed to confirm the human tropism and provide insights into its biological properties.
Keywords: Human; New polyomavirus; Skin.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



References
-
- An P, Saenz Robles MT, Pipas JM, 2012. Large T antigens of polyomaviruses: amazing molecular machines. Annu. Rev. Microbiol 66, 213–236. - PubMed
-
- Azzi A, Fanci R, Bosi A, Ciappi S, Zakrzewska K, de Santis R, Laszlo D, Guidi S, Saccardi R, Vannucchi AM, et al., 1994. Monitoring of polyomavirus BK viruria in bone marrow transplantation patients by DNA hybridization assay and by polymerase chain reaction: an approach to assess the relationship between BK viruria and hemorrhagic cystitis. Bone Marrow Transplant. 14, 235–240. - PubMed
-
- Buck CB, Van Doorslaer K, Peretti A, Geoghegan EM, Tisza MJ, An P, Katz JP, Pipas JM, McBride AA, Camus AC, McDermott AJ, Dill JA, Delwart E, Ng TF, Farkas K, Austin C, Kraberger S, Davison W, Pastrana DV, Varsani A, 2016. The ancient evolutionary history of polyomaviruses. PLoS Pathog. 12, e1005574. - PMC - PubMed
-
- Calvignac-Spencer S, Feltkamp MC, Daugherty MD, Moens U, Ramqvist T, Johne R, Ehlers B, 2016. A taxonomy update for the family Polyomaviridae. Arch. Virol 161, 1739–1750. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

