Functional Architectures of Local and Distal Regulation of Gene Expression in Multiple Human Tissues
- PMID: 28343628
- PMCID: PMC5384099
- DOI: 10.1016/j.ajhg.2017.03.002
Functional Architectures of Local and Distal Regulation of Gene Expression in Multiple Human Tissues
Abstract
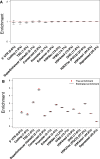
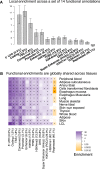
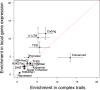
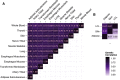
Genetic variants that modulate gene expression levels play an important role in the etiology of human diseases and complex traits. Although large-scale eQTL mapping studies routinely identify many local eQTLs, the molecular mechanisms by which genetic variants regulate expression remain unclear, particularly for distal eQTLs, which these studies are not well powered to detect. Here, we leveraged all variants (not just those that pass stringent significance thresholds) to analyze the functional architecture of local and distal regulation of gene expression in 15 human tissues by employing an extension of stratified LD-score regression that produces robust results in simulations. The top enriched functional categories in local regulation of peripheral-blood gene expression included coding regions (11.41×), conserved regions (4.67×), and four histone marks (p < 5 × 10-5 for all enrichments); local enrichments were similar across the 15 tissues. We also observed substantial enrichments for distal regulation of peripheral-blood gene expression: coding regions (4.47×), conserved regions (4.51×), and two histone marks (p < 3 × 10-7 for all enrichments). Analyses of the genetic correlation of gene expression across tissues confirmed that local regulation of gene expression is largely shared across tissues but that distal regulation is highly tissue specific. Our results elucidate the functional components of the genetic architecture of local and distal regulation of gene expression.
Keywords: eQTLs; functional annotation; gene expression regulation; heritability.
Copyright © 2017 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

