Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis
- PMID: 28344082
- PMCID: PMC5387670
- DOI: 10.1016/j.molcel.2017.02.017
Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis
Abstract
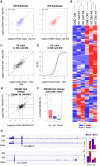
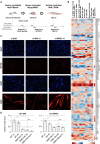
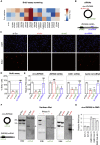
Circular RNAs (circRNAs) constitute a family of transcripts with unique structures and still largely unknown functions. Their biogenesis, which proceeds via a back-splicing reaction, is fairly well characterized, whereas their role in the modulation of physiologically relevant processes is still unclear. Here we performed expression profiling of circRNAs during in vitro differentiation of murine and human myoblasts, and we identified conserved species regulated in myogenesis and altered in Duchenne muscular dystrophy. A high-content functional genomic screen allowed the study of their functional role in muscle differentiation. One of them, circ-ZNF609, resulted in specifically controlling myoblast proliferation. Circ-ZNF609 contains an open reading frame spanning from the start codon, in common with the linear transcript, and terminating at an in-frame STOP codon, created upon circularization. Circ-ZNF609 is associated with heavy polysomes, and it is translated into a protein in a splicing-dependent and cap-independent manner, providing an example of a protein-coding circRNA in eukaryotes.
Keywords: DMD; cap independent; circRNA; circular RNA; muscle differentiation; myogenesis; non-coding RNA; proliferation; translation.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




Comment in
-
RNA: Translated circular RNAs.Nat Rev Genet. 2017 May;18(5):272-273. doi: 10.1038/nrg.2017.27. Epub 2017 Apr 3. Nat Rev Genet. 2017. PMID: 28366936 No abstract available.
References
-
- Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. - PubMed
-
- Ballard D.H., Brown C.M. First Edition. Prentice Hall; 1982. Computer Vision.
-
- Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

