Comparative Transcriptome Reveals Benzenoid Biosynthesis Regulation as Inducer of Floral Scent in the Woody Plant Prunus mume
- PMID: 28344586
- PMCID: PMC5345196
- DOI: 10.3389/fpls.2017.00319
Comparative Transcriptome Reveals Benzenoid Biosynthesis Regulation as Inducer of Floral Scent in the Woody Plant Prunus mume
Abstract
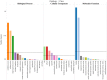
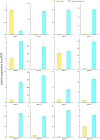
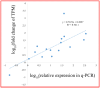
Mei (Prunus mume) is a peculiar woody ornamental plant famous for its inviting fragrance in winter. However, in this valuable plant, the mechanism behind floral volatile development remains poorly defined. Therefore, to explore the floral scent formation, a comparative transcriptome was conducted in order to identify the global transcripts specifying flower buds and blooming flowers of P. mume. Differentially expressed genes were identified between the two different stages showing great discrepancy in floral volatile production. Moreover, according to the expression specificity among the organs (stem, root, fruit, leaf), we summarized one gene cluster regulating the benzenoid floral scent. Significant gene changes were observed in accordance with the formation of benzenoid, thus pointing the pivotal roles of genes as well as cytochrome-P450s and short chain dehydrogenases in the benzenoid biosynthetic process. Further, transcription factors like EMISSION OF BENZENOID I and ODORANT I performed the same expression pattern suggesting key roles in the management of the downstream genes. Taken together, these data provide potential novel anchors for the benzenoid pathway, and the insight for the floral scent induction and regulation mechanism in woody plants.
Keywords: Mei; benzenoid biosynthesis; floral scent; transcription factors; transcriptome.
Figures







References
-
- Audic S., Claverie J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995. - PubMed
-
- Chandran A. K., Lee G. S., Yoo Y. H., Yoon U. H., Ahn B. O., Yun D. W., et al. (2016). Functional classification of rice flanking sequence tagged genes using MapMan terms and global understanding on metabolic and regulatory pathways affected by dxr mutant having defects in light response. Rice (N Y). 9, 17. 10.1186/s12284-016-0089-2 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

