The Dockstore: enabling modular, community-focused sharing of Docker-based genomics tools and workflows
- PMID: 28344774
- PMCID: PMC5333608
- DOI: 10.12688/f1000research.10137.1
The Dockstore: enabling modular, community-focused sharing of Docker-based genomics tools and workflows
Abstract
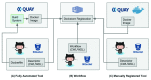
As genomic datasets continue to grow, the feasibility of downloading data to a local organization and running analysis on a traditional compute environment is becoming increasingly problematic. Current large-scale projects, such as the ICGC PanCancer Analysis of Whole Genomes (PCAWG), the Data Platform for the U.S. Precision Medicine Initiative, and the NIH Big Data to Knowledge Center for Translational Genomics, are using cloud-based infrastructure to both host and perform analysis across large data sets. In PCAWG, over 5,800 whole human genomes were aligned and variant called across 14 cloud and HPC environments; the processed data was then made available on the cloud for further analysis and sharing. If run locally, an operation at this scale would have monopolized a typical academic data centre for many months, and would have presented major challenges for data storage and distribution. However, this scale is increasingly typical for genomics projects and necessitates a rethink of how analytical tools are packaged and moved to the data. For PCAWG, we embraced the use of highly portable Docker images for encapsulating and sharing complex alignment and variant calling workflows across highly variable environments. While successful, this endeavor revealed a limitation in Docker containers, namely the lack of a standardized way to describe and execute the tools encapsulated inside the container. As a result, we created the Dockstore ( https://dockstore.org), a project that brings together Docker images with standardized, machine-readable ways of describing and running the tools contained within. This service greatly improves the sharing and reuse of genomics tools and promotes interoperability with similar projects through emerging web service standards developed by the Global Alliance for Genomics and Health (GA4GH).
Keywords: Docker; big data; bioinformatics; cloud; containers; genomics.
Conflict of interest statement
Competing interests: No competing interests were disclosed.
Figures



References
-
- Dirk M: Docker: lightweight linux containers for consistent development and deployment. Linux Journal. 2014;239:2 Reference Source
-
- Mark L, Siu LL, Rehm HL, et al. : All the World's a Stage: Facilitating Discovery Science and Improved Cancer Care through the Global Alliance for Genomics and Health. Cancer Discov. 2015;5(11):1133–1136. 10.1158/2159-8290.CD-15-0821 - DOI - PubMed
-
- Barry L: Oauth web authorization protocol. IEEE Internet Computing. 2012;16(1):74– 77 10.1109/MIC.2012.11 - DOI
-
- Thomas FR: Architectural styles and the design of network-based software architectures.University of California, Irvine.2000. Reference Source
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

