WT1 expression in breast cancer disrupts the epithelial/mesenchymal balance of tumour cells and correlates with the metabolic response to docetaxel
- PMID: 28345629
- PMCID: PMC5366898
- DOI: 10.1038/srep45255
WT1 expression in breast cancer disrupts the epithelial/mesenchymal balance of tumour cells and correlates with the metabolic response to docetaxel
Abstract
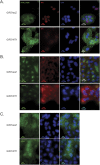
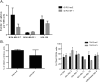
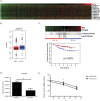
WT1 is a transcription factor which regulates the epithelial-mesenchymal balance during embryonic development and, if mutated, can lead to the formation of Wilms' tumour, the most common paediatric kidney cancer. Its expression has also been reported in several adult tumour types, including breast cancer, and usually correlates with poor outcome. However, published data is inconsistent and the role of WT1 in this malignancy remains unclear. Here we provide a complete study of WT1 expression across different breast cancer subtypes as well as isoform specific expression analysis. Using in vitro cell lines, clinical samples and publicly available gene expression datasets, we demonstrate that WT1 plays a role in regulating the epithelial-mesenchymal balance of breast cancer cells and that WT1-expressing tumours are mainly associated with a mesenchymal phenotype. WT1 gene expression also correlates with CYP3A4 levels and is associated with poorer response to taxane treatment. Our work is the first to demonstrate that the known association between WT1 expression in breast cancer and poor prognosis is potentially due to cancer-related epithelial-to-mesenchymal transition (EMT) and poor chemotherapy response.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

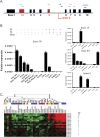
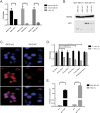
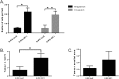
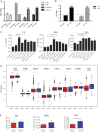
Similar articles
-
Wilms' tumor protein induces an epithelial-mesenchymal hybrid differentiation state in clear cell renal cell carcinoma.PLoS One. 2014 Jul 15;9(7):e102041. doi: 10.1371/journal.pone.0102041. eCollection 2014. PLoS One. 2014. PMID: 25025131 Free PMC article.
-
Classification of a frameshift/extended and a stop mutation in WT1 as gain-of-function mutations that activate cell cycle genes and promote Wilms tumour cell proliferation.Hum Mol Genet. 2014 Aug 1;23(15):3958-74. doi: 10.1093/hmg/ddu111. Epub 2014 Mar 11. Hum Mol Genet. 2014. PMID: 24619359 Free PMC article.
-
Insulin-like growth factor I regulates the expression of isoforms of Wilms' tumor 1 gene in breast cancer.Tumori. 2013 Nov-Dec;99(6):715-22. doi: 10.1177/030089161309900612. Tumori. 2013. PMID: 24503796
-
The role of WT1 in breast cancer: clinical implications, biological effects and molecular mechanism.Int J Biol Sci. 2020 Feb 24;16(8):1474-1480. doi: 10.7150/ijbs.39958. eCollection 2020. Int J Biol Sci. 2020. PMID: 32210734 Free PMC article. Review.
-
The role of Wt1 in regulating mesenchyme in cancer, development, and tissue homeostasis.Trends Genet. 2012 Oct;28(10):515-24. doi: 10.1016/j.tig.2012.04.004. Epub 2012 Jun 1. Trends Genet. 2012. PMID: 22658804 Review.
Cited by
-
RNA Sequencing Identifies WT1 Overexpression as a Predictor of Poor Outcomes in Acute Myeloid Leukemia.Cancers (Basel). 2025 May 29;17(11):1818. doi: 10.3390/cancers17111818. Cancers (Basel). 2025. PMID: 40507300 Free PMC article.
-
Anti-Tumor Immunity to Patient-Derived Breast Cancer Cells by Vaccination with Interferon-Alpha-Conditioned Dendritic Cells (IFN-DC).Vaccines (Basel). 2024 Sep 17;12(9):1058. doi: 10.3390/vaccines12091058. Vaccines (Basel). 2024. PMID: 39340087 Free PMC article.
-
Biologically informed NeuralODEs for genome-wide regulatory dynamics.Genome Biol. 2024 May 21;25(1):127. doi: 10.1186/s13059-024-03264-0. Genome Biol. 2024. PMID: 38773638 Free PMC article.
-
Truncated WT1 Protein Isoform Expression Is Increased in MCF-7 Cells with Long-Term Estrogen Depletion.Int J Breast Cancer. 2021 Nov 20;2021:6282514. doi: 10.1155/2021/6282514. eCollection 2021. Int J Breast Cancer. 2021. PMID: 34845427 Free PMC article.
-
The prognostic significance of human ovarian aging-related signature in breast cancer after surgery: A multicohort study.Front Immunol. 2023 Mar 7;14:1139797. doi: 10.3389/fimmu.2023.1139797. eCollection 2023. Front Immunol. 2023. PMID: 36960071 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

