Smoking-Dependent Distal-to-Proximal Repatterning of the Adult Human Small Airway Epithelium
- PMID: 28345955
- PMCID: PMC5549864
- DOI: 10.1164/rccm.201608-1672OC
Smoking-Dependent Distal-to-Proximal Repatterning of the Adult Human Small Airway Epithelium
Abstract
Rationale: Small airways are the primary site of pathologic changes in chronic obstructive pulmonary disease (COPD), the major smoking-induced lung disorder.
Objectives: On the basis of the concept of proximal-distal patterning that determines regional specialization of the airway epithelium during lung development, we hypothesized that a similar program operates in the adult human lung being altered by smoking, leading to decreased regional identity of the small airway epithelium (SAE).
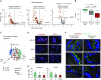
Methods: The proximal and distal airway signatures were identified by comparing the transcriptomes of large and small airway epithelium samples obtained by bronchoscopy from healthy nonsmokers. The expression of these signatures was evaluated in the SAE of healthy smokers and smokers with COPD compared with that of healthy nonsmokers. The capacity of airway basal stem cells (BCs) to maintain region-associated phenotypes was evaluated using the air-liquid interface model.
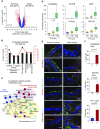
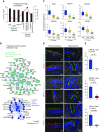
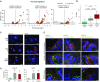
Measurements and main results: The distal and proximal airway signatures, containing 134 and 233 genes, respectively, were identified. These signatures included known developmental regulators of airway patterning, as well as novel regulators such as epidermal growth factor receptor, which was associated with the proximal airway phenotype. In the SAE of smokers with COPD, there was a dramatic smoking-dependent loss of the regional transcriptome identity with concomitant proximalization. This repatterning phenotype was reproduced by stimulating SAE BCs with epidermal growth factor, which was up-regulated in the SAE of smokers, during differentiation of SAE BCs in vitro.
Conclusions: Smoking-induced global distal-to-proximal reprogramming of the SAE represents a novel pathologic feature of COPD and is mediated by exaggerated epidermal growth factor/epidermal growth factor receptor signaling in SAE BCs.
Keywords: basal cells; chronic obstructive pulmonary disease; differentiation; stem cells; transcriptome.
Figures

Comment in
-
Epithelial-Mesenchymal Transition: A Necessary New Therapeutic Target in Chronic Obstructive Pulmonary Disease?Am J Respir Crit Care Med. 2017 Aug 1;196(3):393-394. doi: 10.1164/rccm.201704-0771LE. Am J Respir Crit Care Med. 2017. PMID: 28640653 No abstract available.
-
Reply: Epithelial-Mesenchymal Transition: A Necessary New Therapeutic Target in Chronic Obstructive Pulmonary Disease?Am J Respir Crit Care Med. 2017 Aug 1;196(3):394-395. doi: 10.1164/rccm.201705-0981LE. Am J Respir Crit Care Med. 2017. PMID: 28640654 Free PMC article. No abstract available.
-
More Than Meets the Eye: Cigarette Smoke Induces Genomic Changes in the Small Airway Epithelium Independent of Histologic Changes.Am J Respir Crit Care Med. 2017 Aug 1;196(3):260-262. doi: 10.1164/rccm.201704-0665ED. Am J Respir Crit Care Med. 2017. PMID: 28762786 Free PMC article. No abstract available.
References
-
- Hajj R, Baranek T, Le Naour R, Lesimple P, Puchelle E, Coraux C. Basal cells of the human adult airway surface epithelium retain transit-amplifying cell properties. Stem Cells. 2007;25:139–148. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

