De Novo Assembly, Annotation, and Characterization of Root Transcriptomes of Three Caladium Cultivars with a Focus on Necrotrophic Pathogen Resistance/Defense-Related Genes
- PMID: 28346370
- PMCID: PMC5412298
- DOI: 10.3390/ijms18040712
De Novo Assembly, Annotation, and Characterization of Root Transcriptomes of Three Caladium Cultivars with a Focus on Necrotrophic Pathogen Resistance/Defense-Related Genes
Abstract
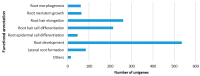
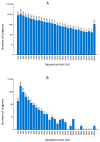
Roots are vital to plant survival and crop yield, yet few efforts have been made to characterize the expressed genes in the roots of non-model plants (root transcriptomes). This study was conducted to sequence, assemble, annotate, and characterize the root transcriptomes of three caladium cultivars (Caladium × hortulanum) using RNA-Seq. The caladium cultivars used in this study have different levels of resistance to Pythiummyriotylum, the most damaging necrotrophic pathogen to caladium roots. Forty-six to 61 million clean reads were obtained for each caladium root transcriptome. De novo assembly of the reads resulted in approximately 130,000 unigenes. Based on bioinformatic analysis, 71,825 (52.3%) caladium unigenes were annotated for putative functions, 48,417 (67.4%) and 31,417 (72.7%) were assigned to Gene Ontology (GO) and Clusters of Orthologous Groups (COG), respectively, and 46,406 (64.6%) unigenes were assigned to 128 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. A total of 4518 distinct unigenes were observed only in Pythium-resistant "Candidum" roots, of which 98 seemed to be involved in disease resistance and defense responses. In addition, 28,837 simple sequence repeat sites and 44,628 single nucleotide polymorphism sites were identified among the three caladium cultivars. These root transcriptome data will be valuable for further genetic improvement of caladium and related aroids.
Keywords: Caladium; RNA-Seq; aroid; de novo assembly; defense-related gene; disease resistance gene; root transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Cao Z., Mclaughlin M., Deng Z. Interspecific genome size and chromosome number variation shed new light on species classification and evolution in caladium. J. Am. Soc. Hortic. Sci. 2014;139:449–459.
-
- Gong L., Deng Z. Development and characterization of microsatellite markers for caladiums (Caladium Vent.) Plant Breed. 2011;130:591–595. doi: 10.1111/j.1439-0523.2011.01863.x. - DOI
-
- Deng Z., Goktepe F., Harbaugh B.K., Hu J.G. Assessment of genetic diversity and relationships among caladium cultivars and species using molecular markers. J. Am. Soc. Hortic. Sci. 2007;132:219–229.
-
- Deng Z., Harbaugh B.K., Kelly R.O., Seijo T., McGovern R.J. Pythium root rot resistance in caladium cultivars. HortScience. 2005;40:549–552.
-
- Deng Z., Harbaugh B.K., Kelly R.O., Seijo T., McGovern R.J. Screening for resistance to pythium root rot among twenty-three caladium cultivars. HortTechnology. 2005;15:631–634.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

