Hepatic cyclooxygenase-2 overexpression induced spontaneous hepatocellular carcinoma formation in mice
- PMID: 28346420
- PMCID: PMC5543258
- DOI: 10.1038/onc.2017.73
Hepatic cyclooxygenase-2 overexpression induced spontaneous hepatocellular carcinoma formation in mice
Abstract
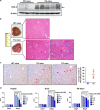
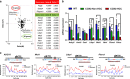
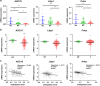
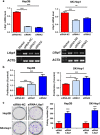
Cyclooxygenase (COX)-2 is upregulated in hepatocellular carcinoma (HCC). However, the direct causative effect of COX-2 in spontaneous HCC formation remains unknown. We thus investigate the role and molecular pathogenesis of COX-2 in HCC by using liver-specific COX-2 transgenic (TG) mice. We found spontaneous HCC formation with elevated inflammatory infiltrates and neovessels in male TG mice (3/21, 14.3%), but not in any of male WT mice (0/19). Reduced representation bisulfite sequencing (RRBS) and gene expression microarrays were performed in the HCC tumor and non-HCC liver tissues to investigate the molecular mechanisms of COX-2-driven HCC. By RRBS, DNA promoter hypermethylation was identified in HCC from TG mice. Induction of promoter hypermethylation was associated with reduced tet methylcytosine dioxygenase 1 (TET1) expression by COX-2. TET1 could catalyze the conversion of 5-methylcytosine into 5-hydroxymethylcytosine (5hmC) and prevents DNA hypermethylation. In keeping with this, loss of 5hmC was demonstrated in COX-2-induced HCC. Consistently, COX-2 overexpression in human HCC cell lines could reduce both TET1 expression and 5hmc levels. Integrative analyses of DNA methylation and gene expression profiles further identified significantly downregulated genes including LTBP1, ADCY5 and PRKCZ by promoter methylation in COX-2-induced HCC. Reduced expression of LTBP1, ADCY5 and PRKCZ by promoter hypermethylation was further validated in human HCCs. Bio-functional investigation revealed that LTBP1 inhibited cell proliferation in HCC cell lines, suggesting its potential role as a tumor suppressor in HCC. Gene expression microarrays revealed that signaling cascades (AKT (protein kinase B), STK33 (Serine/Threonine kinase 33) and MTOR (mechanistic target of rapamycin) pathways) were enriched in COX-2-induced HCC. In conclusion, this study demonstrated for the first time that enhanced COX-2 expression in hepatocytes is sufficient to induce HCC through inducing promoter hypermethylation by reducing TET1, silencing tumor-suppressive genes and activating key oncogenic pathways. Inhibition of COX-2 represents a mechanism-based target for HCC prevention.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Gupta S, Srivastava M, Ahmad N, Bostwick DG, Mukhtar H. Over-expression of cyclooxygenase-2 in human prostate adenocarcinoma. Prostate 2000; 42: 73–78. - PubMed
-
- Singh B, Berry JA, Shoher A, Ramakrishnan V, Lucci A. COX-2 overexpression increases motility and invasion of breast cancer cells. Int J Oncol 2005; 26: 1393–1399. - PubMed
-
- Lim HY, Joo HJ, Choi JH, Yi JW, Yang MS, Cho DY et al. Increased expression of cyclooxygenase-2 protein in human gastric carcinoma. Clin Cancer Res 2000; 6: 519–525. - PubMed
-
- Yip-Schneider MT, Barnard DS, Billings SD, Cheng L, Heilman DK, Lin A et al. Cyclooxygenase-2 expression in human pancreatic adenocarcinomas. Carcinogenesis 2000; 21: 139–146. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

