Altering the threshold of an excitable signal transduction network changes cell migratory modes
- PMID: 28346441
- PMCID: PMC5394931
- DOI: 10.1038/ncb3495
Altering the threshold of an excitable signal transduction network changes cell migratory modes
Abstract
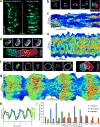
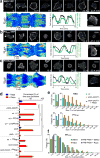
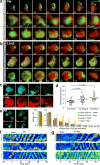
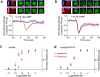
The diverse migratory modes displayed by different cell types are generally believed to be idiosyncratic. Here we show that the migratory behaviour of Dictyostelium was switched from amoeboid to keratocyte-like and oscillatory modes by synthetically decreasing phosphatidylinositol-4,5-bisphosphate levels or increasing Ras/Rap-related activities. The perturbations at these key nodes of an excitable signal transduction network initiated a causal chain of events: the threshold for network activation was lowered, the speed and range of propagating waves of signal transduction activity increased, actin-driven cellular protrusions expanded and, consequently, the cell migratory mode transitions ensued. Conversely, innately keratocyte-like and oscillatory cells were promptly converted to amoeboid by inhibition of Ras effectors with restoration of directed migration. We use computational analysis to explain how thresholds control cell migration and discuss the architecture of the signal transduction network that gives rise to excitability.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. - PubMed
-
- Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. - PubMed
-
- Reichman-Fried M, Minina S, Raz E. Autonomous modes of behavior in primordial germ cell migration. Dev Cell. 2004;6:589–596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

