Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection
- PMID: 28346446
- PMCID: PMC7457382
- DOI: 10.1038/nmicrobiol.2017.37
Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection
Abstract
During the last few decades, the global incidence of dengue virus (DENV) has increased dramatically, and it is now endemic in more than 100 countries. To establish a productive infection in humans, DENV uses different strategies to inhibit or avoid the host innate immune system. Several DENV proteins have been shown to strategically target crucial components of the type I interferon system. Here, we report that the DENV NS2B protease cofactor targets the DNA sensor cyclic GMP-AMP synthase (cGAS) for lysosomal degradation to avoid the detection of mitochondrial DNA during infection. Such degradation subsequently results in the inhibition of type I interferon production in the infected cell. Our data demonstrate a mechanism by which cGAS senses cellular damage upon DENV infection.
Conflict of interest statement
Competing interests
The authors declare no competing financial interests.
Figures



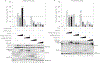
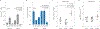

Comment in
-
Viral pathogenesis: Dengue virus takes on cGAS.Nat Microbiol. 2017 Mar 27;2:17050. doi: 10.1038/nmicrobiol.2017.50. Nat Microbiol. 2017. PMID: 28346454 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

