Aberrant neuronal activity-induced signaling and gene expression in a mouse model of RASopathy
- PMID: 28346493
- PMCID: PMC5386306
- DOI: 10.1371/journal.pgen.1006684
Aberrant neuronal activity-induced signaling and gene expression in a mouse model of RASopathy
Erratum in
-
Correction: Aberrant neuronal activity-induced signaling and gene expression in a mouse model of RASopathy.PLoS Genet. 2017 Jun 16;13(6):e1006843. doi: 10.1371/journal.pgen.1006843. eCollection 2017 Jun. PLoS Genet. 2017. PMID: 28622382 Free PMC article.
Abstract
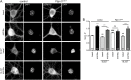
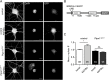
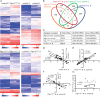
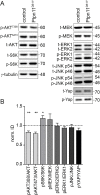
Noonan syndrome (NS) is characterized by reduced growth, craniofacial abnormalities, congenital heart defects, and variable cognitive deficits. NS belongs to the RASopathies, genetic conditions linked to mutations in components and regulators of the Ras signaling pathway. Approximately 50% of NS cases are caused by mutations in PTPN11. However, the molecular mechanisms underlying cognitive impairments in NS patients are still poorly understood. Here, we report the generation and characterization of a new conditional mouse strain that expresses the overactive Ptpn11D61Y allele only in the forebrain. Unlike mice with a global expression of this mutation, this strain is viable and without severe systemic phenotype, but shows lower exploratory activity and reduced memory specificity, which is in line with a causal role of disturbed neuronal Ptpn11 signaling in the development of NS-linked cognitive deficits. To explore the underlying mechanisms we investigated the neuronal activity-regulated Ras signaling in brains and neuronal cultures derived from this model. We observed an altered surface expression and trafficking of synaptic glutamate receptors, which are crucial for hippocampal neuronal plasticity. Furthermore, we show that the neuronal activity-induced ERK signaling, as well as the consecutive regulation of gene expression are strongly perturbed. Microarray-based hippocampal gene expression profiling revealed profound differences in the basal state and upon stimulation of neuronal activity. The neuronal activity-dependent gene regulation was strongly attenuated in Ptpn11D61Y neurons. In silico analysis of functional networks revealed changes in the cellular signaling beyond the dysregulation of Ras/MAPK signaling that is nearly exclusively discussed in the context of NS at present. Importantly, changes in PI3K/AKT/mTOR and JAK/STAT signaling were experimentally confirmed. In summary, this study uncovers aberrant neuronal activity-induced signaling and regulation of gene expression in Ptpn11D61Y mice and suggests that these deficits contribute to the pathophysiology of cognitive impairments in NS.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
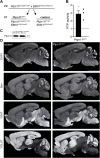
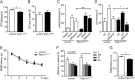
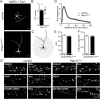

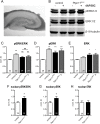

References
-
- Cleghon V, Feldmann P, Ghiglione C, Copeland TD, Perrimon N, et al. (1998) Opposing actions of CSW and RasGAP modulate the strength of Torso RTK signaling in the Drosophila terminal pathway. Mol Cell 2: 719–727. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

