Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c)
- PMID: 28346784
- PMCID: PMC6033327
- DOI: 10.1021/acs.biochem.6b01066
Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c)
Abstract
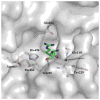
The Mycobacterium tuberculosis (Mtb) serine protease Hip1 (hydrolase important for pathogenesis; Rv2224c) promotes tuberculosis (TB) pathogenesis by impairing host immune responses through proteolysis of a protein substrate, Mtb GroEL2. The cell surface localization of Hip1 and its immunomodulatory functions make Hip1 a good drug target for new adjunctive immune therapies for TB. Here, we report the crystal structure of Hip1 to a resolution of 2.6 Å and the kinetic studies of the enzyme against model substrates and the protein GroEL2. The structure shows a two-domain protein, one of which contains the catalytic residues that are the signature of a serine protease. Surprisingly, a threonine is located within the active site close enough to hydrogen bond with the catalytic residues Asp463 and His490. Mutation of this residue, Thr466, to alanine established its importance for function. Our studies provide insights into the structure of a member of a novel family of proteases. Knowledge of the Hip1 structure will aid in designing inhibitors that could block Hip1 activity.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



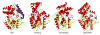

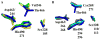

References
-
- Global Tuberculosis Report 2015. World Health Organization (WHO); Geneva, Switzerland: 2015.
-
- Russell DG. Mycobacterium tuberculosis: here today, and here tomorrow. Nat Rev Mol Cell Biol. 2001;2:569–586. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

