Identification and analysis of genomic islands in Burkholderia cenocepacia AU 1054 with emphasis on pathogenicity islands
- PMID: 28347342
- PMCID: PMC5369199
- DOI: 10.1186/s12866-017-0986-6
Identification and analysis of genomic islands in Burkholderia cenocepacia AU 1054 with emphasis on pathogenicity islands
Abstract
Background: Genomic islands (GIs) are genomic regions that reveal evidence of horizontal DNA transfer. They can code for many functions and may augment a bacterium's adaptation to its host or environment. GIs have been identified in strain J2315 of Burkholderia cenocepacia, whereas in strain AU 1054 there has been no published works on such regions according to our text mining and keyword search in Medline.
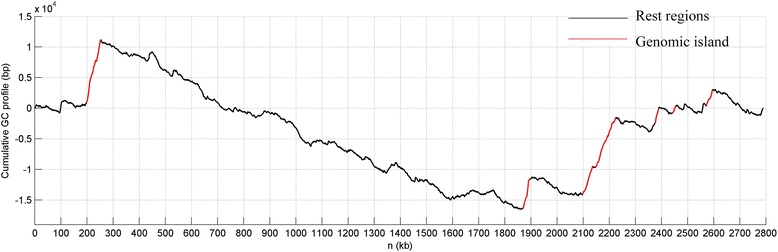
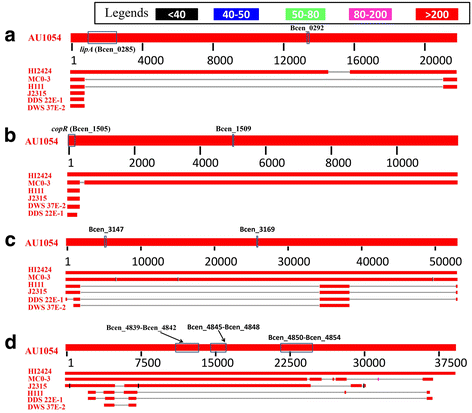
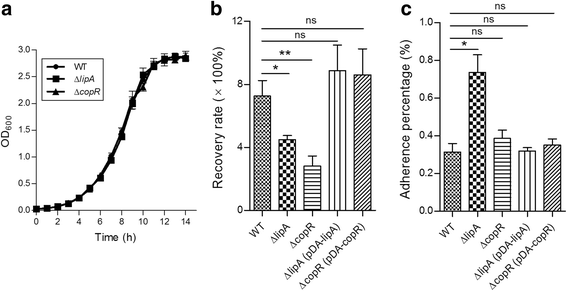
Results: In this study, we identified 21 GIs in AU 1054 by combining two computational tools. Feature analyses suggested that the predictions are highly reliable and hence illustrated the advantage of joint predictions by two independent methods. Based on putative virulence factors, four GIs were further identified as pathogenicity islands (PAIs). Through experiments of gene deletion mutants in live bacteria, two putative PAIs were confirmed, and the virulence factors involved were identified as lipA and copR. The importance of the genes lipA (from PAI 1) and copR (from PAI 2) for bacterial invasion and replication indicates that they are required for the invasive properties of B. cenocepacia and may function as virulence determinants for bacterial pathogenesis and host infection.
Conclusions: This approach of in silico prediction of GIs and subsequent identification of potential virulence factors in the putative island regions with final validation using wet experiments could be used as an effective strategy to rapidly discover novel virulence factors in other bacterial species and strains.
Keywords: B. cenocepacia AU1054; Genomic Island; Pathogenicity Island; Virulence factor.
Figures
References
-
- Regan KH, Bhatt J. Eradication therapy for Burkholderia cepacia complex in people with cystic fibrosis. Cochrane Database Syst Rev. 2014;10:CD009876. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

