Analysis of Shigella flexneri Resistance, Biofilm Formation, and Transcriptional Profile in Response to Bile Salts
- PMID: 28348056
- PMCID: PMC5442615
- DOI: 10.1128/IAI.01067-16
Analysis of Shigella flexneri Resistance, Biofilm Formation, and Transcriptional Profile in Response to Bile Salts
Abstract
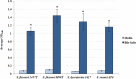
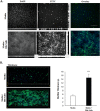
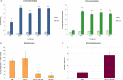
The Shigella species cause millions of cases of watery or bloody diarrhea each year, mostly in children in developing countries. While many aspects of Shigella colonic cell invasion are known, crucial gaps in knowledge regarding how the bacteria survive, transit, and regulate gene expression prior to infection remain. In this study, we define mechanisms of resistance to bile salts and build on previous research highlighting induced virulence in Shigella flexneri strain 2457T following exposure to bile salts. Typical growth patterns were observed within the physiological range of bile salts; however, growth was inhibited at higher concentrations. Interestingly, extended periods of exposure to bile salts led to biofilm formation, a conserved phenotype that we observed among members of the Enterobacteriaceae Characterization of S. flexneri 2457T biofilms determined that both bile salts and glucose were required for formation, dispersion was dependent upon bile salts depletion, and recovered bacteria displayed induced adherence to HT-29 cells. RNA-sequencing analysis verified an important bile salt transcriptional profile in S. flexneri 2457T, including induced drug resistance and virulence gene expression. Finally, functional mutagenesis identified the importance of the AcrAB efflux pump and lipopolysaccharide O-antigen synthesis for bile salt resistance. Our data demonstrate that S. flexneri 2457T employs multiple mechanisms to survive exposure to bile salts, which may have important implications for multidrug resistance. Furthermore, our work confirms that bile salts are important physiological signals to activate S. flexneri 2457T virulence. This work provides insights into how exposure to bile likely regulates Shigella survival and virulence during host transit and subsequent colonic infection.
Keywords: EPS matrix; Escherichia coli; Shigella; bile salts; biofilm; resistance; virulence; virulence genes.
Copyright © 2017 American Society for Microbiology.
Figures





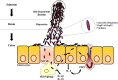
Similar articles
-
Shigella flexneri Adherence Factor Expression in In Vivo-Like Conditions.mSphere. 2019 Nov 13;4(6):e00751-19. doi: 10.1128/mSphere.00751-19. mSphere. 2019. PMID: 31722995 Free PMC article.
-
The Autotransporter IcsA Promotes Shigella flexneri Biofilm Formation in the Presence of Bile Salts.Infect Immun. 2019 Jun 20;87(7):e00861-18. doi: 10.1128/IAI.00861-18. Print 2019 Jul. Infect Immun. 2019. PMID: 30988059 Free PMC article.
-
Shigella flexneri effectors OspE1 and OspE2 mediate induced adherence to the colonic epithelium following bile salts exposure.Mol Microbiol. 2012 Jul;85(1):107-21. doi: 10.1111/j.1365-2958.2012.08092.x. Epub 2012 May 23. Mol Microbiol. 2012. PMID: 22571618 Free PMC article.
-
Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri.Trends Microbiol. 2000 Jan;8(1):17-23. doi: 10.1016/s0966-842x(99)01646-7. Trends Microbiol. 2000. PMID: 10637639 Review.
-
[Molecular genetic studies on virulence-associated chromosomal loci of Shigella flexneri 2 a].Nihon Saikingaku Zasshi. 1993 Nov;48(6):729-38. doi: 10.3412/jsb.48.729. Nihon Saikingaku Zasshi. 1993. PMID: 8309077 Review. Japanese. No abstract available.
Cited by
-
Evaluation of Bile Salts on the Survival and Modulation of Virulence of Aliarcobacter butzleri.Antibiotics (Basel). 2023 Aug 30;12(9):1387. doi: 10.3390/antibiotics12091387. Antibiotics (Basel). 2023. PMID: 37760684 Free PMC article.
-
Genomic, transcriptomic, and phenotypic differences among archetype Shigella flexneri strains of serotypes 2a, 3a, and 6.mSphere. 2023 Dec 20;8(6):e0040823. doi: 10.1128/msphere.00408-23. Epub 2023 Oct 13. mSphere. 2023. PMID: 37830809 Free PMC article.
-
Functional assays to evaluate antibody-mediated responses against Shigella: a review.Front Cell Infect Microbiol. 2023 May 16;13:1171213. doi: 10.3389/fcimb.2023.1171213. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37260708 Free PMC article. Review.
-
Role of the MDR Efflux Pump AcrAB in Epithelial Cell Invasion by Shigella flexneri.Biomolecules. 2023 May 11;13(5):823. doi: 10.3390/biom13050823. Biomolecules. 2023. PMID: 37238693 Free PMC article.
-
Shigella sonnei and Shigella flexneri infection in Caenorhabditis elegans led to species-specific regulatory responses in the host and pathogen.Microb Genom. 2025 Jan;11(1):001339. doi: 10.1099/mgen.0.001339. Microb Genom. 2025. PMID: 39853209 Free PMC article.
References
-
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi:10.1016/S0140-6736(13)60844-2. - DOI - PubMed
-
- Livio S, Strockbine NA, Panchalingam S, Tennant SM, Barry EM, Marohn ME, Antonio M, Hossain A, Mandomando I, Ochieng JB, Oundo JO, Qureshi S, Ramamurthy T, Tamboura B, Adegbola RA, Hossain MJ, Saha D, Sen S, Faruque AS, Alonso PL, Breiman RF, Zaidi AK, Sur D, Sow SO, Berkeley LY, O'Reilly CE, Mintz ED, Biswas K, Cohen D, Farag TH, Nasrin D, Wu Y, Blackwelder WC, Kotloff KL, Nataro JP, Levine MM. 2014. Shigella isolates from the global enteric multicenter study inform vaccine development. Clin Infect Dis 59:933–941. doi:10.1093/cid/ciu468. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

