Structure of human promyeloperoxidase (proMPO) and the role of the propeptide in processing and maturation
- PMID: 28348079
- PMCID: PMC5437232
- DOI: 10.1074/jbc.M117.775031
Structure of human promyeloperoxidase (proMPO) and the role of the propeptide in processing and maturation
Abstract
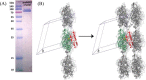
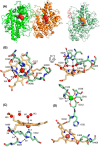
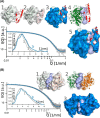
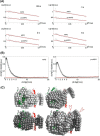
Myeloperoxidase (MPO) is synthesized by neutrophil and monocyte precursor cells and contributes to host defense by mediating microbial killing. Although several steps in MPO biosynthesis and processing have been elucidated, many questions remained, such as the structure-function relationship of monomeric unprocessed proMPO versus the mature dimeric MPO and the functional role of the propeptide. Here we have presented the first and high resolution (at 1.25 Å) crystal structure of proMPO and its solution structure obtained by small-angle X-ray scattering. Promyeloperoxidase hosts five occupied glycosylation sites and six intrachain cystine bridges with Cys-158 of the very flexible N-terminal propeptide being covalently linked to Cys-319 and thereby hindering homodimerization. Furthermore, the structure revealed (i) the binding site of proMPO-processing proconvertase, (ii) the structural motif for subsequent cleavage to the heavy and light chains of mature MPO protomers, and (iii) three covalent bonds between heme and the protein. Studies of the mutants C158A, C319A, and C158A/C319A demonstrated significant differences from the wild-type protein, including diminished enzymatic activity and prevention of export to the Golgi due to prolonged association with the chaperone calnexin. These structural and functional findings provide novel insights into MPO biosynthesis and processing.
Keywords: biosynthesis; crystallography; heme; innate immunity; myeloperoxidase.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
-
- Zamocky M., Jakopitsch C., Furtmüller P. G., Dunand C., and Obinger C. (2008) The peroxidase-cyclooxygenase superfamily: reconstructed evolution of critical enzymes of the innate immune system. Proteins 72, 589–605 - PubMed
-
- Fiedler T. J., Davey C. A., and Fenna R. E. (2000) X-ray crystal structure and characterization of halide binding sites of human myeloperoxidase at 1.8 Å resolution. J. Biol. Chem. 275, 11964–11971 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

