Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain
- PMID: 28348170
- PMCID: PMC5435868
- DOI: 10.1261/rna.059378.116
Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain
Abstract
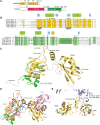
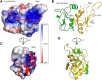
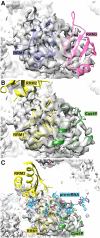
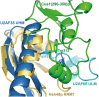
Spliceosomal proteins Hsh49p and Cus1p are components of SF3b, which together with SF3a, Msl1p/Lea1p, Sm proteins, and U2 snRNA, form U2 snRNP, which plays a crucial role in pre-mRNA splicing. Hsh49p, comprising two RRMs, forms a heterodimer with Cus1p. We determined the crystal structures of Saccharomyces cerevisiae full-length Hsh49p as well as its RRM1 in complex with a minimal binding region of Cus1p (residues 290-368). The structures show that the Cus1 fragment binds to the α-helical surface of Hsh49p RRM1, opposite the four-stranded β-sheet, leaving the canonical RNA-binding surface available to bind RNA. Hsh49p binds the 5' end region of U2 snRNA via RRM1. Its affinity is increased in complex with Cus1(290-368)p, partly because an extended RNA-binding surface forms across the protein-protein interface. The Hsh49p RRM1-Cus1(290-368)p structure fits well into cryo-EM density of the Bact spliceosome, corroborating the biological relevance of our crystal structure.
Keywords: RNA binding; RRM; SF3b; U2 snRNP; splicing.
© 2017 van Roon et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures
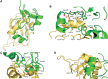
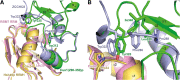


References
-
- Barraud P, Allain FHT. 2013. Solution structure of the two RNA recognition motifs of hnRNP A1 using segmental isotope labeling: how the relative orientation between RRMs influences the nucleic acid binding topology. J Biomol NMR 55: 119–138. - PubMed
-
- Bertram K, Agafonov DE, Liu W-T, Dybkov O, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lührmann R. 2017. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542: 318–323. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
