Live imaging of root-bacteria interactions in a microfluidics setup
- PMID: 28348235
- PMCID: PMC5410799
- DOI: 10.1073/pnas.1618584114
Live imaging of root-bacteria interactions in a microfluidics setup
Abstract
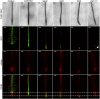
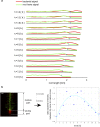
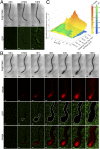
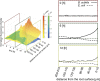
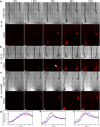
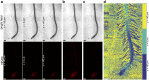
Plant roots play a dominant role in shaping the rhizosphere, the environment in which interaction with diverse microorganisms occurs. Tracking the dynamics of root-microbe interactions at high spatial resolution is currently limited because of methodological intricacy. Here, we describe a microfluidics-based approach enabling direct imaging of root-bacteria interactions in real time. The microfluidic device, which we termed tracking root interactions system (TRIS), consists of nine independent chambers that can be monitored in parallel. The principal assay reported here monitors behavior of fluorescently labeled Bacillus subtilis as it colonizes the root of Arabidopsis thaliana within the TRIS device. Our results show a distinct chemotactic behavior of B. subtilis toward a particular root segment, which we identify as the root elongation zone, followed by rapid colonization of that same segment over the first 6 h of root-bacteria interaction. Using dual inoculation experiments, we further show active exclusion of Escherichia coli cells from the root surface after B. subtilis colonization, suggesting a possible protection mechanism against root pathogens. Furthermore, we assembled a double-channel TRIS device that allows simultaneous tracking of two root systems in one chamber and performed real-time monitoring of bacterial preference between WT and mutant root genotypes. Thus, the TRIS microfluidics device provides unique insights into the microscale microbial ecology of the complex root microenvironment and is, therefore, likely to enhance the current rate of discoveries in this momentous field of research.
Keywords: Bacillus subtilis; TRIS; live-imaging microscopy; microbial community dynamics; root–bacteria interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
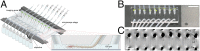
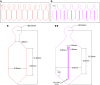

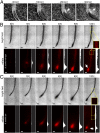
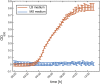

Comment in
-
Shining a light on the dark world of plant root-microbe interactions.Proc Natl Acad Sci U S A. 2017 Apr 25;114(17):4281-4283. doi: 10.1073/pnas.1703800114. Epub 2017 Apr 4. Proc Natl Acad Sci U S A. 2017. PMID: 28377510 Free PMC article. No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

