Bonobos Maintain Immune System Diversity with Three Functional Types of MHC-B
- PMID: 28348269
- PMCID: PMC5469624
- DOI: 10.4049/jimmunol.1601955
Bonobos Maintain Immune System Diversity with Three Functional Types of MHC-B
Abstract
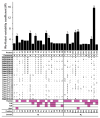
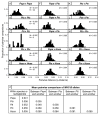
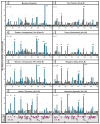
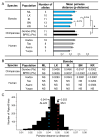
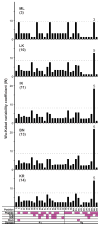
Fast-evolving MHC class I polymorphism serves to diversify NK cell and CD8 T cell responses in individuals, families, and populations. Because only chimpanzee and bonobo have strict orthologs of all HLA class I, their study gives unique perspectives on the human condition. We defined polymorphism of Papa-B, the bonobo ortholog of HLA-B, for six wild bonobo populations. Sequences for Papa-B exon 2 and 3 were determined from the genomic DNA in 255 fecal samples, minimally representing 110 individuals. Twenty-two Papa-B alleles were defined, each encoding a different Papa-B protein. No Papa-B is identical to any chimpanzee Patr-B, human HLA-B, or gorilla Gogo-B. Phylogenetic analysis identified a clade of MHC-B, defined by residues 45-74 of the α1 domain, which is broadly conserved among bonobo, chimpanzee, and gorilla. Bonobo populations have 3-14 Papa-B allotypes. Three Papa-B are in all populations, and they are each of a different functional type: allotypes having the Bw4 epitope recognized by killer cell Ig-like receptors of NK cells, allotypes having the C1 epitope also recognized by killer cell Ig-like receptors, and allotypes having neither epitope. For population Malebo, these three Papa-B are the only Papa-B allotypes. Although small in number, their sequence divergence is such that the nucleotide diversity (mean proportional distance) of Papa-B in Malebo is greater than in the other populations and is also greater than expected for random combinations of three Papa-B Overall, Papa-B has substantially less diversity than Patr-B in chimpanzee subspecies and HLA-B in indigenous human populations, consistent with bonobo having experienced narrower population bottlenecks.
Copyright © 2017 by The American Association of Immunologists, Inc.
Figures





References
-
- Kelley J, Walter L, Trowsdale J. Comparative genomics of major histocompatibility complexes. Immunogenetics. 2005;56:683–695. - PubMed
-
- Brown JH, Jardetzky T, Saper MA, Samraoui B, Bjorkman PJ, Wiley DC. A hypothetical model of the foreign antigen binding site of Class II histocompatibility molecules. Nature. 1988;332:845–850. - PubMed
-
- Villadangos JA. Presentation of antigens by MHC class II molecules: getting the most out of them. Mol Immunol. 2001;38:329–346. - PubMed
-
- Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. - PubMed
-
- Zinkernagel RM, Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974;248:701–702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

