A redox-mediated Kemp eliminase
- PMID: 28348375
- PMCID: PMC5379065
- DOI: 10.1038/ncomms14876
A redox-mediated Kemp eliminase
Abstract
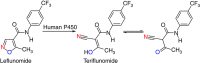
The acid/base-catalysed Kemp elimination of 5-nitro-benzisoxazole forming 2-cyano-4-nitrophenol has long served as a design platform of enzymes with non-natural reactions, providing new mechanistic insights in protein science. Here we describe an alternative concept based on redox catalysis by P450-BM3, leading to the same Kemp product via a fundamentally different mechanism. QM/MM computations show that it involves coordination of the substrate's N-atom to haem-Fe(II) with electron transfer and concomitant N-O heterolysis liberating an intermediate having a nitrogen radical moiety Fe(III)-N· and a phenoxyl anion. Product formation occurs by bond rotation and H-transfer. Two rationally chosen point mutations cause a notable increase in activity. The results shed light on the prevailing mechanistic uncertainties in human P450-catalysed metabolism of the immunomodulatory drug leflunomide, which likewise undergoes redox-mediated Kemp elimination by P450-BM3. Other isoxazole-based pharmaceuticals are probably also metabolized by a redox mechanism. Our work provides a basis for designing future artificial enzymes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Kemp D., Cox D. D. & Paul K. G. Physical organic chemistry of benzisoxazoles. IV. Origins and catalytic nature of the solvent rate acceleration for the decarboxylation of 3-carboxybenzisoxazoles. J. Am. Chem. Soc. 97, 7312–7318 (1975).
-
- Albanese D. C. & Gaggero N. Albumin as a promiscuous biocatalyst in organic synthesis. RSC Adv. 5, 10588–10598 (2015).
-
- Thorn S. N., Daniels R. G., Auditor M.-T. M. & Hilvert D. Large rate accelerations in antibody catalysis by strategic use of haptenic charge. Nature 373, 228–230 (1995). - PubMed
-
- Hollfelder F., Kirby A. J. & Tawfik D. S. Off-the-shelf proteins that rival tailor-made antibodies as catalysts. Nature 383, 60–63 (1996). - PubMed
-
- Kikuchi K., Thorn S. N. & Hilvert D. Albumin-catalyzed proton transfer. J. Am. Chem. Soc. 118, 8184–8185 (1996).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

