Copper-Catalyzed Click Reaction on/in Live Cells
- PMID: 28348729
- PMCID: PMC5365239
- DOI: 10.1039/C6SC02297A
Copper-Catalyzed Click Reaction on/in Live Cells
Abstract
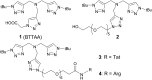
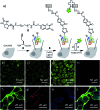
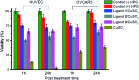
We demonstrated that copper-catalyzed azide-alkyne cycloaddition (CuAAC) reaction could be performed inside live mammalian cells without using a chelating azide. Under optimized conditions, the reaction was performed in human ovary cancer cell line OVCAR5 in which newly synthesized proteins were metabolically modified with homopropargylglycine (HPG). This model system allowed us to estimate the efficiency of the reaction on the cell membranes and in the cytosol using mass spectrometry. We found that the reaction was greatly promoted by a tris(triazolylmethyl)amine CuI ligand tethering a cell-penetrating peptide. Uptake of the ligand, copper, and a biotin-tagged azide in the cells was determined to be 69 ± 2, 163 ± 3 and 1.3 ± 0.1 µM, respectively. After 10 minutes of reaction, the product yields on the membrane and cytosolic proteins were higher than 18% and 0.8%, respectively, while 75% cells remained viable. By reducing the biothiols in the system by scraping or treatment with N-ethylmalemide, the reaction yield on the cytosolic proteins was greatly improved to ~9% and ~14%, respectively, while the yield on the membrane proteins remained unchanged. The results indicate that out of many possibilities, deactivation of the current copper catalysts by biothiols is the major reason for the low yield of CuAAC reaction in the cytosol. Overall, we have improved the efficiency for CuAAC reaction on live cells by 3-fold. Despite the low yielding inside live cells, the products that strongly bind to the intracellular targets can be detected by mass spectrometry. Hence, the in situ CuAAC reaction can be potentially used for screening of cell-specific enzyme inhibitors or biomarkers containing 1,4-substituted 1,2,3-triazoles.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

