Region-specific diversification of the highly virulent serotype 1 Streptococcus pneumoniae
- PMID: 28348812
- PMCID: PMC5320570
- DOI: 10.1099/mgen.0.000027
Region-specific diversification of the highly virulent serotype 1 Streptococcus pneumoniae
Abstract
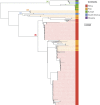
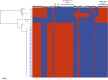
Serotype 1 Streptococcus pneumoniae is a leading cause of invasive pneumococcal disease (IPD) worldwide, with the highest burden in developing countries. We report the whole-genome sequencing analysis of 448 serotype 1 isolates from 27 countries worldwide (including 11 in Africa). The global serotype 1 population shows a strong phylogeographic structure at the continental level, and within Africa there is further region-specific structure. Our results demonstrate that region-specific diversification within Africa has been driven by limited cross-region transfer events, genetic recombination and antimicrobial selective pressure. Clonal replacement of the dominant serotype 1 clones circulating within regions is uncommon; however, here we report on the accessory gene content that has contributed to a rare clonal replacement event of ST3081 with ST618 as the dominant cause of IPD in the Gambia.
Keywords: Africa; Antibiotic Resistance; Genomics; PAGe; Phylogeny; Pneumococcal Disease; Recombination.
Figures






Similar articles
-
Comparative Genomic Analysis and In Vivo Modeling of Streptococcus pneumoniae ST3081 and ST618 Isolates Reveal Key Genetic and Phenotypic Differences Contributing to Clonal Replacement of Serotype 1 in The Gambia.J Infect Dis. 2017 Dec 5;216(10):1318-1327. doi: 10.1093/infdis/jix472. J Infect Dis. 2017. PMID: 28968897 Free PMC article.
-
Serotypes, antimicrobial susceptibility, and molecular epidemiology of invasive and non-invasive Streptococcus pneumoniae isolates in paediatric patients after the introduction of 13-valent conjugate vaccine in a nationwide surveillance study conducted in Japan in 2012-2014.Vaccine. 2016 Jan 2;34(1):67-76. doi: 10.1016/j.vaccine.2015.11.015. Epub 2015 Nov 19. Vaccine. 2016. PMID: 26602268
-
Population structure and drug resistance patterns of emerging non-PCV-13 Streptococcus pneumoniae serotypes 22F, 15A, and 8 isolated from adults in Ontario, Canada.Infect Genet Evol. 2016 Aug;42:1-8. doi: 10.1016/j.meegid.2016.04.007. Epub 2016 Apr 9. Infect Genet Evol. 2016. PMID: 27071529
-
Impact of pneumococcal conjugate vaccine on infections caused by antibiotic-resistant Streptococcus pneumoniae.Clin Microbiol Infect. 2009 Apr;15 Suppl 3:16-20. doi: 10.1111/j.1469-0691.2009.02726.x. Clin Microbiol Infect. 2009. PMID: 19366365 Review.
-
Summary of invasive pneumococcal disease burden among children in the Asia-Pacific region.Vaccine. 2010 Nov 10;28(48):7589-605. doi: 10.1016/j.vaccine.2010.07.053. Epub 2010 Aug 1. Vaccine. 2010. PMID: 20674872 Review.
Cited by
-
Understanding pneumococcal serotype 1 biology through population genomic analysis.BMC Infect Dis. 2016 Nov 8;16(1):649. doi: 10.1186/s12879-016-1987-z. BMC Infect Dis. 2016. PMID: 27821148 Free PMC article.
-
Lower Density and Shorter Duration of Nasopharyngeal Carriage by Pneumococcal Serotype 1 (ST217) May Explain Its Increased Invasiveness over Other Serotypes.mBio. 2020 Dec 8;11(6):e00814-20. doi: 10.1128/mBio.00814-20. mBio. 2020. PMID: 33293378 Free PMC article.
-
Serotype 1 pneumococcus: epidemiology, genomics, and disease mechanisms.Trends Microbiol. 2022 Jun;30(6):581-592. doi: 10.1016/j.tim.2021.11.007. Epub 2021 Dec 20. Trends Microbiol. 2022. PMID: 34949516 Free PMC article. Review.
-
Hospital-based Surveillance for Pediatric Bacterial Meningitis in the Era of the 13-Valent Pneumococcal Conjugate Vaccine in Ghana.Clin Infect Dis. 2019 Sep 5;69(Suppl 2):S89-S96. doi: 10.1093/cid/ciz464. Clin Infect Dis. 2019. PMID: 31505622 Free PMC article.
-
Recombination in Streptococcus pneumoniae Lineages Increase with Carriage Duration and Size of the Polysaccharide Capsule.mBio. 2016 Sep 27;7(5):e01053-16. doi: 10.1128/mBio.01053-16. mBio. 2016. PMID: 27677790 Free PMC article.
References
-
- Antonio M., Hakeem I., Awine T., Secka O., Sankareh K., Nsekpong D., Lahai G., Akisanya A., Egere U., other authors (2008). Seasonality and outbreak of a predominant Streptococcus pneumoniae serotype 1 clone from The Gambia: expansion of ST217 hypervirulent clonal complex in West Africa BMC Microbiol 8198.10.1186/1471-2180-8-198.. - DOI - PMC - PubMed
-
- Blumental S., Moïsi J.C., Roalfe L., Zancolli M., Johnson M., Burbidge P., Borrow R., Yaro S., Mueller J.E., other authors (2015). Streptococcuspneumoniae serotype 1 burden in the African meningitis belt: exploration of functionality in specific antibodies Clin Vaccine Immunol 22404–41210.1128/CVI.00758-14. - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

