Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes
- PMID: 28348841
- PMCID: PMC5320581
- DOI: 10.1099/mgen.0.000043
Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes
Abstract
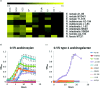
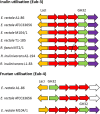
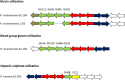
Firmicutes and Bacteroidetes are the predominant bacterial phyla colonizing the healthy human large intestine. Whilst both ferment dietary fibre, genes responsible for this important activity have been analysed only in the Bacteroidetes, with very little known about the Firmicutes. This work investigates the carbohydrate-active enzymes (CAZymes) in a group of Firmicutes, Roseburia spp. and Eubacterium rectale, which play an important role in producing butyrate from dietary carbohydrates and in health maintenance. Genome sequences of 11 strains representing E. rectale and four Roseburia spp. were analysed for carbohydrate-active genes. Following assembly into a pan-genome, core, variable and unique genes were identified. The 1840 CAZyme genes identified in the pan-genome were assigned to 538 orthologous groups, of which only 26 were present in all strains, indicating considerable inter-strain variability. This analysis was used to categorize the 11 strains into four carbohydrate utilization ecotypes (CUEs), which were shown to correspond to utilization of different carbohydrates for growth. Many glycoside hydrolase genes were found linked to genes encoding oligosaccharide transporters and regulatory elements in the genomes of Roseburia spp. and E. rectale, forming distinct polysaccharide utilization loci (PULs). Whilst PULs are also a common feature in Bacteroidetes, key differences were noted in these Firmicutes, including the absence of close homologues of Bacteroides polysaccharide utilization genes, hence we refer to Gram-positive PULs (gpPULs). Most CAZyme genes in the Roseburia/E. rectale group are organized into gpPULs. Variation in gpPULs can explain the high degree of nutritional specialization at the species level within this group.
Keywords: Carbohydrate; Lachnospiraceae; Roseburia; comparative genomics; gut microbiota; obligate anaerobe.
Figures


References
-
- Aminov R. I., Walker A. W., Duncan S. H., Harmsen H. J., Welling G. W., Flint H. J. (2006). Molecular diversity, cultivation, and improved detection by fluorescent in situ hybridization of a dominant group of human gut bacteria related to Roseburia spp. or Eubacterium rectale Appl Environ Microbiol 726371–6376. - PMC - PubMed
-
- Balamurugan R., Rajendiran E., George S., Samuel G. V., Ramakrishna B. S. (2008). Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer J Gastroenterol Hepatol 231298–1303. - PubMed
Data References
-
- Eubacterium rectale T1-815 genome (2015); CVRQ01000001–CVRQ01000090: http://www.ebi.ac.uk/ena/data/view/PRJEB9320.
-
- Roseburia faecis M72/1 genome (2015); CVRR01000001–CVRR01000101: http://www.ebi.ac.uk/ena/data/view/PRJEB9321.
-
- Roseburia inulinivorans L1-83 genome (2015); CVRS01000001–CVRS01000151: http://www.ebi.ac.uk/ena/data/view/PRJEB9322.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

