Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1
- PMID: 28348844
- PMCID: PMC5320584
- DOI: 10.1099/mgen.0.000052
Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1
Erratum in
-
Corrigendum: Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1.Microb Genom. 2019 Jul;5(7):e000280. doi: 10.1099/mgen.0.000280. Microb Genom. 2019. PMID: 31364967 Free PMC article. No abstract available.
Abstract
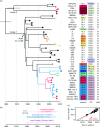
The majority of Acinetobacter baumannii isolates that are multiply, extensively and pan-antibiotic resistant belong to two globally disseminated clones, GC1 and GC2, that were first noticed in the 1970s. Here, we investigated microevolution and phylodynamics within GC1 via analysis of 45 whole-genome sequences, including 23 sequenced for this study. The most recent common ancestor of GC1 arose around 1960 and later diverged into two phylogenetically distinct lineages. In the 1970s, the main lineage acquired the AbaR resistance island, conferring resistance to older antibiotics, via a horizontal gene transfer event. We estimate a mutation rate of ∼5 SNPs genome- 1 year- 1 and detected extensive recombination within GC1 genomes, introducing nucleotide diversity into the population at >20 times the substitution rate (the ratio of SNPs introduced by recombination compared with mutation was 22). The recombination events were non-randomly distributed in the genome and created significant diversity within loci encoding outer surface molecules (including the capsular polysaccharide, the outer core lipooligosaccharide and the outer membrane protein CarO), and spread antimicrobial resistance-conferring mutations affecting the gyrA and parC genes and insertion sequence insertions activating the ampC gene. Both GC1 lineages accumulated resistance to newer antibiotics through various genetic mechanisms, including the acquisition of plasmids and transposons or mutations in chromosomal genes. Our data show that GC1 has diversified into multiple successful extensively antibiotic-resistant subclones that differ in their surface structures. This has important implications for all avenues of control, including epidemiological tracking, antimicrobial therapy and vaccination.
Keywords: Acinetobacter baumannii; antibiotic resistance; capsule; evolution; phylogenomics; recombination.
Figures





References
Data References
-
- Translational Genomics Research Institute(2012). GenBank accession number AMIV01 (Acinetobacter baumannii TG19582, whole-genome shotgun sequence).
-
- Adams, M. D., Goglin, K., Molyneaux, N., Hujer, K. M., Lavender, H., Jamison, J. J., McDonald, I. J., Martin, K. M., Russo, T. & other authors (2008). GenBank accession number CP001172.1 (Acinetobacter baumannii 307-0294, complete genome).
-
- Cerqueira, G., Feldgarden, M., Courvalin, P., Perichon, B., Grillot-Courvalin, C., Clermont, D., Rocha, E., Yoon, E.-J., Nemec, A. & other authors (2013). Short Read Archive accession numbers SRR654309 (Acinetobacter baumannii NIPH 527), SRR654194 (Acinetobacter baumannii NIPH 290) and SRR654201 (Acinetobacter baumannii ANC 4097).
-
- Harkins, D. M., Durkin, A. S., Beck, E., Fedorova, N. B., Kim, M., Onuska, J., Radune, D., DePew, J., Koroleva, G. I. & other authors (2012). Short Read Archive accession numbers SRR387244 (Acinetobacter baumannii OIFC074), SRR387323 (Acinetobacter baumannii Naval-21), SRR387319 (Acinetobacter baumannii Naval-83), SRR387315 (Acinetobacter baumannii Canada-BC1), SRR353953 (Acinetobacter baumannii Canada-BC5) and SRR387296 (Acinetobacter baumannii IS-58).
-
- Adams, M. D., Chan, E. R., Molyneaux, N. & Bonomo, R. A. (2010). GenBank accession numbers ADHA01 (Acinetobacter baumannii AB058, whole-genome shotgun sequence), ADGZ01 (Acinetobacter baumannii AB056, whole-genome shotgun sequence) and ADHB01 (Acinetobacter baumannii AB059, whole-genome shotgun sequence).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

