Metabolic engineering to expand the substrate spectrum of Pseudomonas putida toward sucrose
- PMID: 28349670
- PMCID: PMC5552902
- DOI: 10.1002/mbo3.473
Metabolic engineering to expand the substrate spectrum of Pseudomonas putida toward sucrose
Abstract
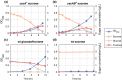
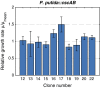
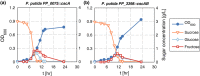
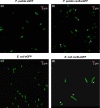
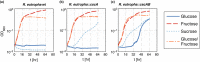
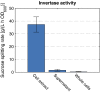
Sucrose is an important disaccharide used as a substrate in many industrial applications. It is a major component of molasses, a cheap by-product of the sugar industry. Unfortunately, not all industrially relevant organisms, among them Pseudomonas putida, are capable of metabolizing sucrose. We chose a metabolic engineering approach to circumvent this blockage and equip P. putida with the activities necessary to consume sucrose. Therefore, we constructed a pair of broad-host range mini-transposons (pSST - sucrose splitting transposon), carrying either cscA, encoding an invertase able to split sucrose into glucose and fructose, or additionally cscB, encoding a sucrose permease. Introduction of cscA was sufficient to convey sucrose consumption and the additional presence of cscB had no further effect, though the sucrose permease was built and localized to the membrane. Sucrose was split extracellularly by the activity of the invertase CscA leaking out of the cell. The transposons were also used to confer sucrose consumption to Cupriavidus necator. Interestingly, in this strain, CscB acted as a glucose transporter, such that C. necator also gained the ability to grow on glucose. Thus, the pSST transposons are functional tools to extend the substrate spectrum of Gram-negative bacterial strains toward sucrose.
Keywords: Pseudomonas putida; metabolic engineering; sucrose metabolism.
© 2017 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures
References
-
- Aparicio, T. , Jensen, S. I. , Nielsen, A. T. , de Lorenzo, V. , & Martínez‐García, E. (2016). The Ssr protein (T1E_1405) from Pseudomonas putida DOT‐T1E enables oligonucleotide‐based recombineering in platform strain P. putida EM42. Biotechnology Journal, 11, 1309–1319. - PubMed
-
- Belda, E. , van Heck, R. G. A. , Lopez‐Sanchez, M. J. , Cruveiller, S. , Barbe, V. , Fraser, C. , … Médigue, C. (2016). The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environmental Microbiology, 18, 3403–3424. - PubMed
-
- Chen, G.‐Q. (2009). A microbial polyhydroxyalkanoates (PHA) based bio‐ and materials industry. Chemical Society Reviews, 38, 2434–2446. - PubMed
-
- de Lorenzo, V. , Eltis, L. , Kessler, B. , & Timmis, K. (1993). Analysis of Pseudomonas gene products using lacIq/Ptrp‐lac plasmids and transposons that confer conditional phenotypes. Gene, 123, 17–24. - PubMed
-
- de Lorenzo, V. , & Timmis, K. (1994). Analysis and construction of stable phenotypes in gram‐negative bacteria with Tn5‐ and Tn10‐derived minitransposons. Methods in Enzymology, 235, 386–405. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

