Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants
- PMID: 28350038
- PMCID: PMC5452131
- DOI: 10.1590/1678-4685-GMB-2016-0106
Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants
Abstract
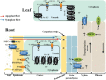
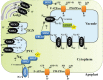
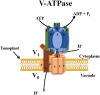
Soil salinity is a major abiotic stress that results in considerable crop yield losses worldwide. However, some plant genotypes show a high tolerance to soil salinity, as they manage to maintain a high K+/Na+ ratio in the cytosol, in contrast to salt stress susceptible genotypes. Although, different plant genotypes show different salt tolerance mechanisms, they all rely on the regulation and function of K+ and Na+ transporters and H+ pumps, which generate the driving force for K+ and Na+ transport. In this review we will introduce salt stress responses in plants and summarize the current knowledge about the most important ion transporters that facilitate intra- and intercellular K+ and Na+ homeostasis in these organisms. We will describe and discuss the regulation and function of the H+-ATPases, H+-PPases, SOS1, HKTs, and NHXs, including the specific tissues where they work and their response to salt stress.
Figures
Similar articles
-
Reassessing the role of ion homeostasis for improving salinity tolerance in crop plants.Physiol Plant. 2021 Apr;171(4):502-519. doi: 10.1111/ppl.13112. Epub 2020 May 5. Physiol Plant. 2021. PMID: 32320060 Review.
-
Regulation of cation transporter genes by the arbuscular mycorrhizal symbiosis in rice plants subjected to salinity suggests improved salt tolerance due to reduced Na(+) root-to-shoot distribution.Mycorrhiza. 2016 Oct;26(7):673-84. doi: 10.1007/s00572-016-0704-5. Epub 2016 Apr 26. Mycorrhiza. 2016. PMID: 27113587
-
A Critical Role of Sodium Flux via the Plasma Membrane Na+/H+ Exchanger SOS1 in the Salt Tolerance of Rice.Plant Physiol. 2019 Jun;180(2):1046-1065. doi: 10.1104/pp.19.00324. Epub 2019 Apr 16. Plant Physiol. 2019. PMID: 30992336 Free PMC article.
-
Ion Transporters and Abiotic Stress Tolerance in Plants.ISRN Mol Biol. 2012 Jun 3;2012:927436. doi: 10.5402/2012/927436. eCollection 2012. ISRN Mol Biol. 2012. PMID: 27398240 Free PMC article. Review.
-
Regulation of ion homeostasis under salt stress.Curr Opin Plant Biol. 2003 Oct;6(5):441-5. doi: 10.1016/s1369-5266(03)00085-2. Curr Opin Plant Biol. 2003. PMID: 12972044 Review.
Cited by
-
Regulation of nitro-oxidative homeostasis: an effective approach to enhance salinity tolerance in plants.Plant Cell Rep. 2024 Jul 15;43(8):193. doi: 10.1007/s00299-024-03275-y. Plant Cell Rep. 2024. PMID: 39008125 Review.
-
TILLING-by-Sequencing+ Reveals the Role of Novel Fatty Acid Desaturases (GmFAD2-2s) in Increasing Soybean Seed Oleic Acid Content.Cells. 2021 May 19;10(5):1245. doi: 10.3390/cells10051245. Cells. 2021. PMID: 34069320 Free PMC article.
-
An Enhanced Interaction of Graft and Exogenous SA on Photosynthesis, Phytohormone, and Transcriptome Analysis in Tomato under Salinity Stress.Int J Mol Sci. 2024 Oct 8;25(19):10799. doi: 10.3390/ijms251910799. Int J Mol Sci. 2024. PMID: 39409129 Free PMC article.
-
Bread Wheat With High Salinity and Sodicity Tolerance.Front Plant Sci. 2019 Oct 22;10:1280. doi: 10.3389/fpls.2019.01280. eCollection 2019. Front Plant Sci. 2019. PMID: 31695711 Free PMC article.
-
Response of Moringa oleifera trees to salinity stress conditions in Tabuk region, Kingdom of Saudi Arabia.Saudi J Biol Sci. 2023 Oct;30(10):103810. doi: 10.1016/j.sjbs.2023.103810. Epub 2023 Sep 14. Saudi J Biol Sci. 2023. PMID: 37766885 Free PMC article.
References
-
- Adams E, Shin R. Transport, signaling, and homeostasis of potassium and sodium in plants. J Integr Plant Biol. 2014;56:231–249. - PubMed
-
- Aharon GS, Apse MP, Duan S, Hua X, Blumwald E. Characterization of a family of vacuolar Na+/H+ antiporters in Arabidopsis thaliana . Plant Soil. 2003;253:245–256.
-
- Alexandersson E, Saalbach G, Larsson C, Kjellbom P. Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol. 2004;45:1543–1556. - PubMed
-
- Ali R, Brett CL, Mukherjee S, Rao R. Inhibition of sodium/proton exchange by a Rab-GTPase-activating protein regulates endosomal traffic in yeast. J Biol Chem. 2004;279:4498–4506. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

