Animal-like prostaglandins in marine microalgae
- PMID: 28350392
- PMCID: PMC5520147
- DOI: 10.1038/ismej.2017.27
Animal-like prostaglandins in marine microalgae
Abstract
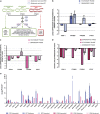
Diatoms are among the most successful primary producers in ocean and freshwater environments. Deriving from a secondary endosymbiotic event, diatoms have a mixed genome containing bacterial, animal and plant genes encoding for metabolic pathways that may account for their evolutionary success. Studying the transcriptomes of two strains of the diatom Skeletonema marinoi, we report, for the first time in microalgae, an active animal-like prostaglandin pathway that is differentially expressed in the two strains. Prostaglandins are hormone-like mediators in many physiological and pathological processes in mammals, playing a pivotal role in inflammatory responses. They are also present in macroalgae and invertebrates, where they act as defense and communication mediators. The occurrence of animal-like prostaglandins in unicellular photosynthetic eukaryotes opens up new intriguing perspectives on the evolution and role of these molecules in the marine environment as possible mediators in cell-to-cell signaling, eventually influencing population dynamics in the plankton.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Gerecht A, Romano G, Ianora A, d’Ippolito G, Cutignano A, Fontana A. (2011). Plasticity of oxylipin metabolism among clones of the marine diatom skeletonema marinoi (bacillariophyceae)1. J Phycol 47: 1050–1056. - PubMed
-
- d’Ippolito G, Tucci S, Cutignano A, Romano G, Cimino G, Miralto A et al. (2004). The role of complex lipids in the synthesis of bioactive aldehydes of the marine diatom Skeletonema costatum. Biochim Biophys Acta 1686: 100–107. - PubMed
-
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA et al. (2014). The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol 12: e1001889. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

