Diversification of spatiotemporal expression and copy number variation of the echinoid hbox12/pmar1/micro1 multigene family
- PMID: 28350855
- PMCID: PMC5370098
- DOI: 10.1371/journal.pone.0174404
Diversification of spatiotemporal expression and copy number variation of the echinoid hbox12/pmar1/micro1 multigene family
Abstract
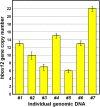
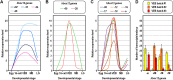
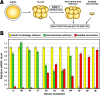
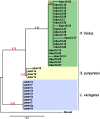
Changes occurring during evolution in the cis-regulatory landscapes of individual members of multigene families might impart diversification in their spatiotemporal expression and function. The archetypal member of the echinoid hbox12/pmar1/micro1 family is hbox12-a, a homeobox-containing gene expressed exclusively by dorsal blastomeres, where it governs the dorsal/ventral gene regulatory network during embryogenesis of the sea urchin Paracentrotus lividus. Here we describe the inventory of the hbox12/pmar1/micro1 genes in P. lividus, highlighting that gene copy number variation occurs across individual sea urchins of the same species. We show that the various hbox12/pmar1/micro1 genes group into three subfamilies according to their spatiotemporal expression, which ranges from broad transcription throughout development to transient expression in either the animal hemisphere or micromeres of the early embryo. Interestingly, the promoter regions of those genes showing comparable expression patterns are highly similar, while differing from those of the other subfamilies. Strikingly, phylogenetic analysis suggests that the hbox12/pmar1/micro1 genes are species-specific, exhibiting extensive divergence in their noncoding, but not in their coding, sequences across three distinct sea urchin species. In spite of this, two micromere-specific genes of P. lividus possess a TCF/LEF-binding motif in a similar position, and their transcription relies on Wnt/β-catenin signaling, similar to the pmar1 and micro1 genes, which in other sea urchin species are involved in micromere specification. Altogether, our findings suggest that the hbox12/pmar1/micro1 gene family evolved rather rapidly, generating paralogs whose cis-regulatory sequences diverged following multiple rounds of duplication from a common ancestor.
Conflict of interest statement
Figures

Similar articles
-
Transient activation of the micro1 homeobox gene family in the sea urchin ( Hemicentrotus pulcherrimus) micromere.Dev Genes Evol. 2002 Feb;212(1):1-10. doi: 10.1007/s00427-001-0202-3. Epub 2002 Jan 23. Dev Genes Evol. 2002. PMID: 11875651
-
pmar1/phb homeobox genes and the evolution of the double-negative gate for endomesoderm specification in echinoderms.Development. 2020 Feb 26;147(4):dev182139. doi: 10.1242/dev.182139. Development. 2020. PMID: 32001441
-
A regulatory gene network that directs micromere specification in the sea urchin embryo.Dev Biol. 2002 Jun 1;246(1):209-28. doi: 10.1006/dbio.2002.0627. Dev Biol. 2002. PMID: 12027443
-
Homeobox genes and sea urchin development.Int J Dev Biol. 2000;44(6):637-43. Int J Dev Biol. 2000. PMID: 11061427 Review.
-
Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus).Dev Biol. 2006 Dec 1;300(1):35-48. doi: 10.1016/j.ydbio.2006.08.012. Epub 2006 Aug 10. Dev Biol. 2006. PMID: 16997294 Review.
Cited by
-
Transcription of microRNAs is regulated by developmental signaling pathways and transcription factors.Front Cell Dev Biol. 2024 Apr 24;12:1356589. doi: 10.3389/fcell.2024.1356589. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38721525 Free PMC article.
-
Inducible and reversible inhibition of miRNA-mediated gene repression in vivo.Elife. 2021 Aug 31;10:e70948. doi: 10.7554/eLife.70948. Elife. 2021. PMID: 34463618 Free PMC article.
-
Analysis of the P. lividus sea urchin genome highlights contrasting trends of genomic and regulatory evolution in deuterostomes.Cell Genom. 2023 Apr 5;3(4):100295. doi: 10.1016/j.xgen.2023.100295. eCollection 2023 Apr 12. Cell Genom. 2023. PMID: 37082140 Free PMC article.
-
Antioxidant, Pro-Survival and Pro-Regenerative Effects of Conditioned Medium from Wharton's Jelly Mesenchymal Stem Cells on Developing Zebrafish Embryos.Int J Mol Sci. 2023 Aug 25;24(17):13191. doi: 10.3390/ijms241713191. Int J Mol Sci. 2023. PMID: 37685998 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

