MicroRNAs Establish Uniform Traits during the Architecture of Vertebrate Embryos
- PMID: 28350988
- PMCID: PMC5404386
- DOI: 10.1016/j.devcel.2017.02.021
MicroRNAs Establish Uniform Traits during the Architecture of Vertebrate Embryos
Abstract
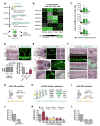
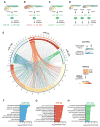
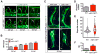
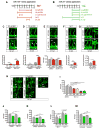
Proper functioning of an organism requires cells and tissues to behave in uniform, well-organized ways. How this optimum of phenotypes is achieved during the development of vertebrates is unclear. Here, we carried out a multi-faceted and single-cell resolution screen of zebrafish embryonic blood vessels upon mutagenesis of single and multi-gene microRNA (miRNA) families. We found that embryos lacking particular miRNA-dependent signaling pathways develop a vascular trait similar to wild-type, but with a profound increase in phenotypic heterogeneity. Aberrant trait variance in miRNA mutant embryos uniquely sensitizes their vascular system to environmental perturbations. We discovered a previously unrecognized role for specific vertebrate miRNAs to protect tissue development against phenotypic variability. This discovery marks an important advance in our comprehension of how miRNAs function in the development of higher organisms.
Keywords: cardiovascular development; endothelial cells; environmental stress; miR-139; miR-223; miR-24; phenotypic variability; robustness; target gene networks; zebrafish mutants.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Comment in
-
MicroRNAs Make a Difference in Cardiovascular Robustness.Dev Cell. 2017 Mar 27;40(6):515-516. doi: 10.1016/j.devcel.2017.03.009. Dev Cell. 2017. PMID: 28350982 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

