Tissue and time specific expression pattern of interferon regulated genes in the chicken
- PMID: 28351377
- PMCID: PMC5371264
- DOI: 10.1186/s12864-017-3641-6
Tissue and time specific expression pattern of interferon regulated genes in the chicken
Abstract
Background: Type I interferons are major players against viral infections and mediate their function by the induction of Interferon regulated genes (IRGs). Recently, it became obvious that these cytokines have a multitude of additional functions. Due to the unique features of the chickens' immune system, available data from mouse models are not easily transferable; hence we performed an extensive analysis of chicken IRGs.
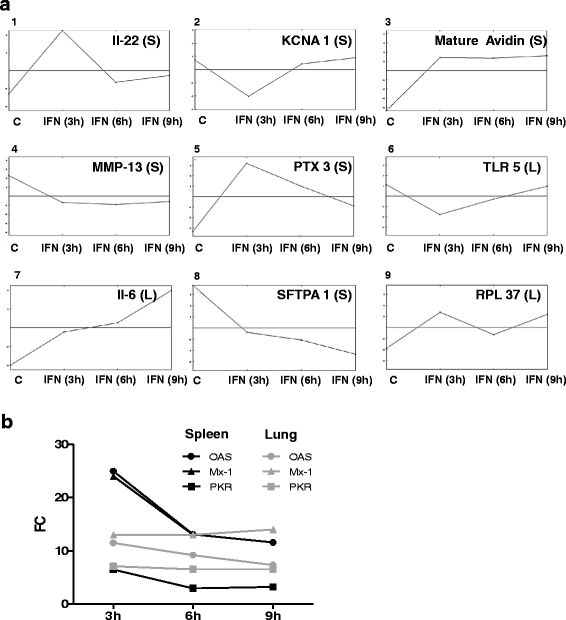
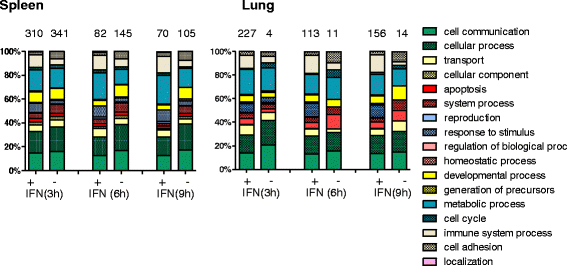
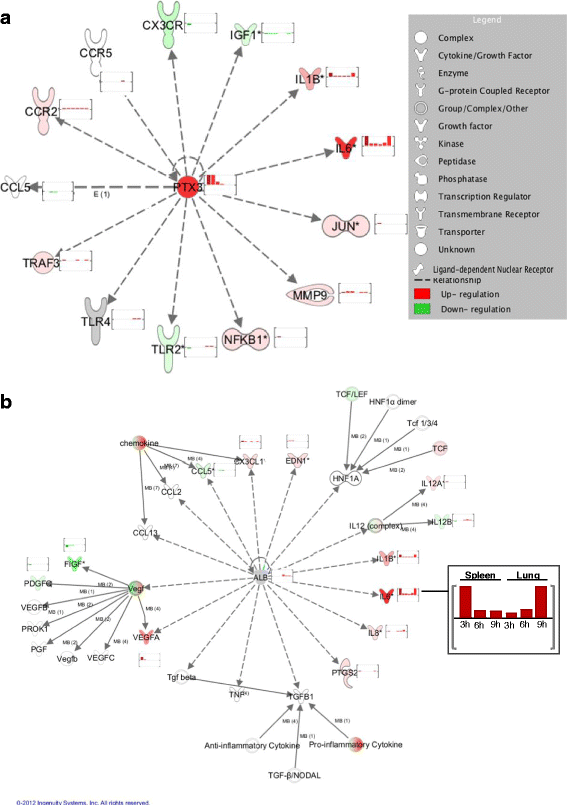
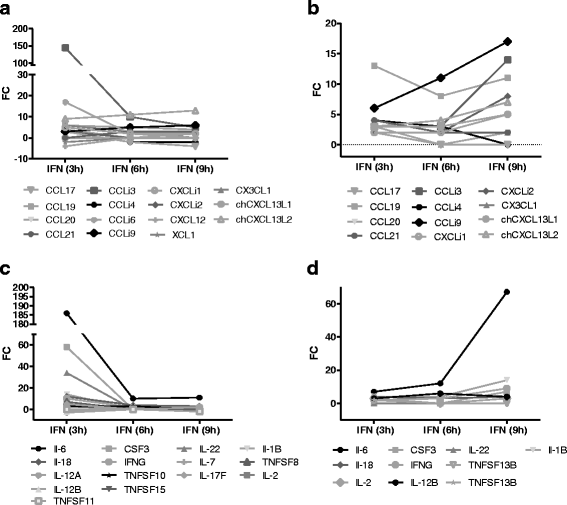
Results: A broad database search for homologues to described mammalian IRGs (common IRGs, cIRGs) was combined with a transcriptome analysis of spleen and lung at different time points after application of IFNα. To apply physiological amounts of IFN, half-life of IFN in the chicken was determined. Interestingly, the calculated 36 min are considerably shorter than the ones obtained for human and mouse. Microarray analysis revealed many additional IRGs (newly identified IRGs; nIRGs) and network analysis for selected IRGs showed a broad interaction of nIRGs among each other and with cIRGs. We found that IRGs exhibit a highly tissue and time specific expression pattern as expression quality and quantity differed strongly between spleen and lung and over time. While in the spleen for many affected genes changes in RNA abundance peaked already after 3 h, an increasing or plateau-like regulation after 3, 6 and 9 h was observed in the lung.
Conclusions: The induction or suppression of IRGs in chickens is both tissue and time specific and beside known antiviral mechanisms type I IFN induces many additional cellular functions. We confirmed many known IRGs and established a multitude of so far undescribed ones, thus providing a large database for future research on antiviral mechanisms and additional IFN functions in non-mammalian species.
Keywords: Chicken; Expression profile; Half-life; IFNα; IRG.
Figures




Similar articles
-
RNA sequencing for global gene expression associated with muscle growth in a single male modern broiler line compared to a foundational Barred Plymouth Rock chicken line.BMC Genomics. 2017 Jan 13;18(1):82. doi: 10.1186/s12864-016-3471-y. BMC Genomics. 2017. PMID: 28086790 Free PMC article.
-
Interferon regulated gene (IRG) expression-signature in a mouse model of chikungunya virus neurovirulence.J Neurovirol. 2017 Dec;23(6):886-902. doi: 10.1007/s13365-017-0583-3. Epub 2017 Oct 24. J Neurovirol. 2017. PMID: 29067635
-
Genome-wide DNA methylome variation in two genetically distinct chicken lines using MethylC-seq.BMC Genomics. 2015 Oct 23;16:851. doi: 10.1186/s12864-015-2098-8. BMC Genomics. 2015. PMID: 26497311 Free PMC article.
-
Regulation of class II MHC gene expression by interferons: insights into the mechanism of action of interferon (review).Anticancer Res. 1988 Nov-Dec;8(6):1153-61. Anticancer Res. 1988. PMID: 2464333 Review.
-
Systems biology of interferon responses.J Interferon Cytokine Res. 2011 Jan;31(1):5-11. doi: 10.1089/jir.2010.0126. J Interferon Cytokine Res. 2011. PMID: 21226606 Review.
Cited by
-
Cholesterol-25-hydroxylase Is a Chicken ISG That Restricts ALV-J Infection by Producing 25-hydroxycholesterol.Viruses. 2019 May 30;11(6):498. doi: 10.3390/v11060498. Viruses. 2019. PMID: 31151272 Free PMC article.
-
The Long Pentraxin PTX3 Is of Major Importance Among Acute Phase Proteins in Chickens.Front Immunol. 2019 Feb 1;10:124. doi: 10.3389/fimmu.2019.00124. eCollection 2019. Front Immunol. 2019. PMID: 30774632 Free PMC article.
-
Transcriptome Analysis in Spleen Reveals Differential Regulation of Response to Newcastle Disease Virus in Two Chicken Lines.Sci Rep. 2018 Jan 19;8(1):1278. doi: 10.1038/s41598-018-19754-8. Sci Rep. 2018. PMID: 29352240 Free PMC article.
-
Interferon Signaling in Chickens Plays a Crucial Role in Inhibiting Influenza Replication in DF1 Cells.Microorganisms. 2022 Jan 10;10(1):133. doi: 10.3390/microorganisms10010133. Microorganisms. 2022. PMID: 35056582 Free PMC article.
-
Cyclic Guanosine Monophosphate-Adenosine Monophosphate Synthase (cGAS), a Multifaceted Platform of Intracellular DNA Sensing.Front Immunol. 2021 Feb 23;12:637399. doi: 10.3389/fimmu.2021.637399. eCollection 2021. Front Immunol. 2021. PMID: 33708225 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

