Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial
- PMID: 28351427
- PMCID: PMC5371199
- DOI: 10.1186/s12916-017-0834-4
Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial
Abstract
Background: RIFAQUIN was a tuberculosis chemotherapy trial in southern Africa including regimens with high-dose rifapentine with moxifloxacin. Here, the application of whole-genome sequencing (WGS) is evaluated within RIFAQUIN for identifying new infections in treated patients as either relapses or reinfections. WGS is further compared with mycobacterial interspersed repetitive units-variable number tandem repeats (MIRU-VNTR) typing. This is the first report of WGS being used to evaluate new infections in a completed clinical trial for which all treatment and epidemiological data are available for analysis.
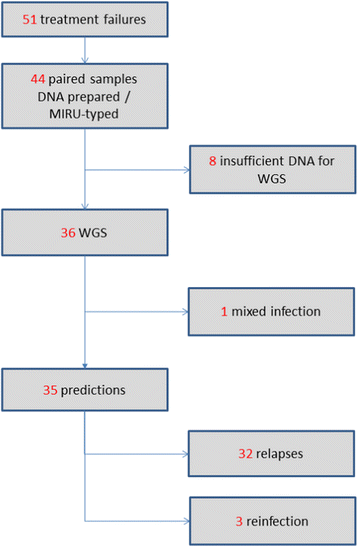
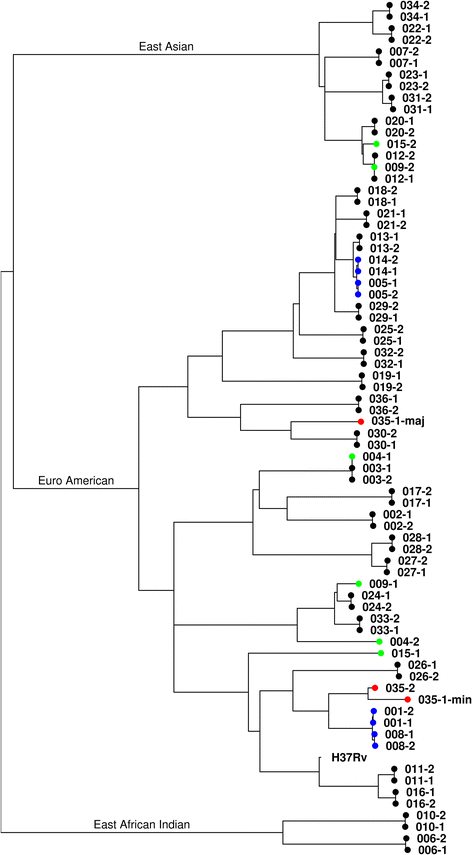
Methods: DNA from 36 paired samples of Mycobacterium tuberculosis cultured from patients before and after treatment was typed using 24-loci MIRU-VNTR, in silico spoligotyping and WGS. Following WGS, the sequences were mapped against the reference strain H37Rv, the single-nucleotide polymorphism (SNP) differences between pairs were identified, and a phylogenetic reconstruction was performed.
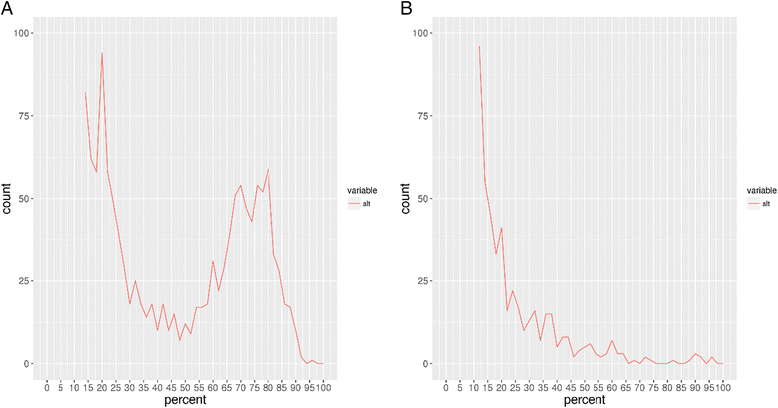
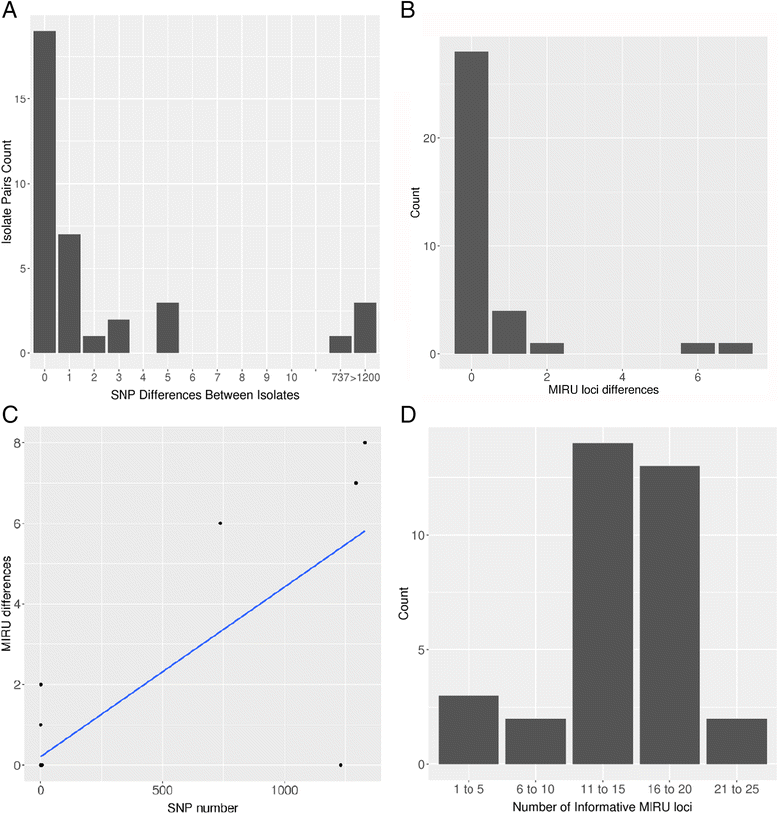
Results: WGS indicated that 32 of the paired samples had a very low number of SNP differences (0-5; likely relapses). One pair had an intermediate number of SNP differences, and was likely the result of a mixed infection with a pre-treatment minor genotype that was highly related to the post-treatment genotype; this was reclassified as a relapse, in contrast to the MIRU-VNTR result. The remaining three pairs had very high SNP differences (>750; likely reinfections).
Conclusions: WGS and MIRU-VNTR both similarly differentiated relapses and reinfections, but WGS provided significant extra information. The low proportion of reinfections seen suggests that in standard chemotherapy trials with up to 24 months of follow-up, typing the strains brings little benefit to an analysis of the trial outcome in terms of differentiating relapse and reinfection. However, there is a benefit to using WGS as compared to MIRU-VNTR in terms of the additional genotype information obtained, in particular for defining the presence of mixed infections and the potential to identify known and novel drug-resistance markers.
Keywords: Clinical trial; Tuberculosis; Whole genome sequencing.
Figures
Similar articles
-
Genomic sequencing is required for identification of tuberculosis transmission in Hawaii.BMC Infect Dis. 2018 Dec 3;18(1):608. doi: 10.1186/s12879-018-3502-1. BMC Infect Dis. 2018. PMID: 30509214 Free PMC article.
-
Strain typing of Mycobacterium tuberculosis isolates from Korea by mycobacterial interspersed repetitive units-variable number of tandem repeats.Korean J Lab Med. 2009 Aug;29(4):314-9. doi: 10.3343/kjlm.2009.29.4.314. Korean J Lab Med. 2009. PMID: 19726893
-
Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis.J Clin Microbiol. 2008 Apr;46(4):1398-406. doi: 10.1128/JCM.02089-07. Epub 2008 Jan 30. J Clin Microbiol. 2008. PMID: 18234864 Free PMC article.
-
The Evolution of Strain Typing in the Mycobacterium tuberculosis Complex.Adv Exp Med Biol. 2017;1019:43-78. doi: 10.1007/978-3-319-64371-7_3. Adv Exp Med Biol. 2017. PMID: 29116629 Review.
-
Contact investigations for outbreaks of Mycobacterium tuberculosis: advances through whole genome sequencing.Clin Microbiol Infect. 2013 Sep;19(9):796-802. doi: 10.1111/1469-0691.12183. Epub 2013 Feb 21. Clin Microbiol Infect. 2013. PMID: 23432709 Review.
Cited by
-
CD4+ T cells are homeostatic regulators during Mtb reinfection.bioRxiv [Preprint]. 2023 Dec 21:2023.12.20.572669. doi: 10.1101/2023.12.20.572669. bioRxiv. 2023. PMID: 38187598 Free PMC article. Preprint.
-
Simplified Model to Survey Tuberculosis Transmission in Countries Without Systematic Molecular Epidemiology Programs.Emerg Infect Dis. 2019 Mar;25(3):507-514. doi: 10.3201/eid2503.181593. Emerg Infect Dis. 2019. PMID: 30789134 Free PMC article.
-
Considerations for biomarker-targeted intervention strategies for tuberculosis disease prevention.Tuberculosis (Edinb). 2018 Mar;109:61-68. doi: 10.1016/j.tube.2017.11.009. Epub 2017 Nov 22. Tuberculosis (Edinb). 2018. PMID: 29559122 Free PMC article.
-
Methods for Detecting Mycobacterial Mixed Strain Infections-A Systematic Review.Front Genet. 2020 Dec 21;11:600692. doi: 10.3389/fgene.2020.600692. eCollection 2020. Front Genet. 2020. PMID: 33408740 Free PMC article.
-
Baseline and acquired resistance to bedaquiline, linezolid and pretomanid, and impact on treatment outcomes in four tuberculosis clinical trials containing pretomanid.PLOS Glob Public Health. 2023 Oct 18;3(10):e0002283. doi: 10.1371/journal.pgph.0002283. eCollection 2023. PLOS Glob Public Health. 2023. PMID: 37851685 Free PMC article.
References
-
- Fox W, Ellard GA, Mitchison DA. Studies on the treatment of tuberculosis undertaken by the British Medical Research Council tuberculosis units, 1946-1986, with relevant subsequent publications. Int J Tuberc Lung Dis. 1999;3(10 Suppl 2):S231–279. - PubMed
-
- Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44(12):4498–510. doi: 10.1128/JCM.01392-06. - DOI - PMC - PubMed
-
- Das S, Chan SL, Allen BW, Mitchison DA, Lowrie DB. Application of DNA fingerprinting with IS986 to sequential mycobacterial isolates obtained from pulmonary tuberculosis patients in Hong Kong before, during and after short-course chemotherapy. Tuber Lung Dis. 1993;74(1):47–51. doi: 10.1016/0962-8479(93)90068-9. - DOI - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

