Structural and biophysical analyses of the skeletal dihydropyridine receptor β subunit β1a reveal critical roles of domain interactions for stability
- PMID: 28351836
- PMCID: PMC5437245
- DOI: 10.1074/jbc.M116.763896
Structural and biophysical analyses of the skeletal dihydropyridine receptor β subunit β1a reveal critical roles of domain interactions for stability
Abstract
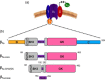
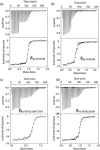
Excitation-contraction (EC) coupling in skeletal muscle requires a physical interaction between the voltage-gated calcium channel dihydropyridine receptor (DHPR) and the ryanodine receptor Ca2+ release channel. Although the exact molecular mechanism that initiates skeletal EC coupling is unresolved, it is clear that both the α1 and β subunits of DHPR are essential for this process. Here, we employed a series of techniques, including size-exclusion chromatography-multi-angle light scattering, differential scanning fluorimetry, and isothermal calorimetry, to characterize various biophysical properties of the skeletal DHPR β subunit β1a Removal of the intrinsically disordered N and C termini and the hook region of β1a prevented oligomerization, allowing for its structural determination by X-ray crystallography. The structure had a topology similar to that of previously determined β isoforms, which consist of SH3 and guanylate kinase domains. However, transition melting temperatures derived from the differential scanning fluorimetry experiments indicated a significant difference in stability of ∼2-3 °C between the β1a and β2a constructs, and the addition of the DHPR α1s I-II loop (α-interaction domain) peptide stabilized both β isoforms by ∼6-8 °C. Similar to other β isoforms, β1a bound with nanomolar affinity to the α-interaction domain, but binding affinities were influenced by amino acid substitutions in the adjacent SH3 domain. These results suggest that intramolecular interactions between the SH3 and guanylate kinase domains play a role in the stability of β1a while also providing a conduit for allosteric signaling events.
Keywords: X-ray crystallography; dihydropyridine receptor (DHPR); excitation-contraction coupling (E-C coupling); protein structure; skeletal muscle.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

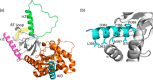



References
-
- Obermair G. J., Tuluc P., and Flucher B. E. (2008) Auxiliary Ca2+ channel subunits: lessons learned from muscle. Curr. Opin. Pharmacol. 8, 311–318 - PubMed
-
- Coronado R., Ahern C. A., Sheridan D. C., Cheng W., Carbonneau L., and Bhattacharya D. (2004) Functional equivalence of dihydropyridine receptor α1S and β1a subunits in triggering excitation-contraction coupling in skeletal muscle. Biol. Res. 37, 565–575 - PubMed
-
- Wu J., Yan Z., Li Z., Yan C., Lu S., Dong M., and Yan N. (2015) Structure of the voltage-gated calcium channel Cav1.1 complex. Science 350, aad2395. - PubMed
-
- Wu J., Yan Z., Li Z., Qian X., Lu S., Dong M., Zhou Q., and Yan N. (2016) Structure of the voltage-gated calcium channel Cav1.1 at 3.6 A resolution. Nature 537, 191–196 - PubMed
-
- Chen Y. H., Li M. H., Zhang Y., He L. L., Yamada Y., Fitzmaurice A., Shen Y., Zhang H., Tong L., and Yang J. (2004) Structural basis of the α1-β subunit interaction of voltage-gated Ca2+ channels. Nature 429, 675–680 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

