Genetic Heterogeneity in Therapy-Naïve Synchronous Primary Breast Cancers and Their Metastases
- PMID: 28351929
- PMCID: PMC5768571
- DOI: 10.1158/1078-0432.CCR-16-3115
Genetic Heterogeneity in Therapy-Naïve Synchronous Primary Breast Cancers and Their Metastases
Abstract
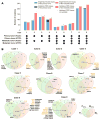
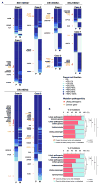
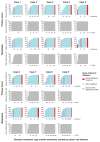
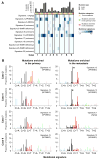
Purpose: Paired primary breast cancers and metachronous metastases after adjuvant treatment are reported to differ in their clonal composition and genetic alterations, but it is unclear whether these differences stem from the selective pressures of the metastatic process, the systemic therapies, or both. We sought to define the repertoire of genetic alterations in breast cancer patients with de novo metastatic disease who had not received local or systemic therapy.Experimental Design: Up to two anatomically distinct core biopsies of primary breast cancers and synchronous distant metastases from nine patients who presented with metastatic disease were subjected to high-depth whole-exome sequencing. Mutations, copy number alterations and their cancer cell fractions, and mutation signatures were defined using state-of-the-art bioinformatics methods. All mutations identified were validated with orthogonal methods.Results: Genomic differences were observed between primary and metastatic deposits, with a median of 60% (range 6%-95%) of shared somatic mutations. Although mutations in known driver genes including TP53, PIK3CA, and GATA3 were preferentially clonal in both sites, primary breast cancers and their synchronous metastases displayed spatial intratumor heterogeneity. Likely pathogenic mutations affecting epithelial-to-mesenchymal transition-related genes, including SMAD4, TCF7L2, and TCF4 (ITF2), were found to be restricted to or enriched in the metastatic lesions. Mutational signatures of trunk mutations differed from those of mutations enriched in the primary tumor or the metastasis in six cases.Conclusions: Synchronous primary breast cancers and metastases differ in their repertoire of somatic genetic alterations even in the absence of systemic therapy. Mutational signature shifts might contribute to spatial intratumor genetic heterogeneity. Clin Cancer Res; 23(15); 4402-15. ©2017 AACR.
©2017 American Association for Cancer Research.
Figures


References
-
- Andre F, Slimane K, Bachelot T, Dunant A, Namer M, Barrelier A, et al. Breast cancer with synchronous metastases: trends in survival during a 14-year period. J Clin Oncol. 2004;22:3302–8. - PubMed
-
- Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. - PubMed
-
- Klein CA. Selection and adaptation during metastatic cancer progression. Nature. 2013;501:365–72. - PubMed
-
- Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

