PKCα diffusion and translocation are independent of an intact cytoskeleton
- PMID: 28352102
- PMCID: PMC5428563
- DOI: 10.1038/s41598-017-00560-7
PKCα diffusion and translocation are independent of an intact cytoskeleton
Abstract
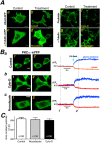
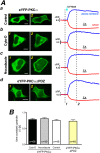
Translocation of cytosolic cPKC to the plasma membrane is a key event in their activation process but its exact nature is still unclear with particular dispute whether sole diffusion or additional active transport along the cell's cytoskeleton contributes to cPKC's dynamics. This was addressed by analyzing the recruitment behavior of PKCα while manipulating the cytoskeleton. Photolytic Ca2+ uncaging allowed us to quantify the kinetics of PKCα redistribution to the plasma membrane when fused to monomeric, dimeric and tetrameric fluorescence proteins. Results indicated that translocation kinetics were modulated by the state of oligomerization as expected for varying Stokes' radii of the participating proteins. Following depolymerization of the microtubules and the actin filaments we found that Ca2+ induced membrane accumulation of PKCα was independent of the filamentous state of the cytoskeleton. Fusion of PKCα to the photo-convertible fluorescent protein Dendra2 enabled the investigation of PKCα-cytoskeleton interactions under resting conditions. Redistribution following spatially restricted photoconversion showed that the mobility of the fusion protein was independent of the state of the cytoskeleton. Our data demonstrated that in living cells neither actin filaments nor microtubules contribute to PKCα's cytosolic mobility or Ca2+-induced translocation to the plasma membrane. Instead translocation is a solely diffusion-driven process.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Simultaneous visualization of the translocation of protein kinase Calpha-green fluorescent protein hybrids and intracellular calcium concentrations.Biochem J. 1999 Jan 15;337 ( Pt 2)(Pt 2):211-8. Biochem J. 1999. PMID: 9882617 Free PMC article.
-
Analysis of the subcellular distribution of protein kinase Calpha using PKC-GFP fusion proteins.Exp Cell Res. 2000 Jul 10;258(1):204-14. doi: 10.1006/excr.2000.4925. Exp Cell Res. 2000. PMID: 10912802
-
PKCalpha: a versatile key for decoding the cellular calcium toolkit.J Cell Biol. 2006 Aug 14;174(4):521-33. doi: 10.1083/jcb.200604033. Epub 2006 Aug 7. J Cell Biol. 2006. PMID: 16893971 Free PMC article.
-
PKCalpha translocation is microtubule-dependent in passaged smooth muscle cells.Acta Physiol Scand. 2000 Oct;170(2):87-97. doi: 10.1046/j.1365-201x.2000.00755.x. Acta Physiol Scand. 2000. PMID: 11114946
-
New directions for fluorescent speckle microscopy.Curr Biol. 2002 Sep 17;12(18):R633-40. doi: 10.1016/s0960-9822(02)01139-9. Curr Biol. 2002. PMID: 12372272 Review.
Cited by
-
Phospho-substrate profiling of Epac-dependent protein kinase C activity.Mol Cell Biochem. 2019 Jun;456(1-2):167-178. doi: 10.1007/s11010-019-03502-1. Epub 2019 Feb 9. Mol Cell Biochem. 2019. PMID: 30739223 Free PMC article.
-
mTORC2 controls the activity of PKC and Akt by phosphorylating a conserved TOR interaction motif.Sci Signal. 2021 Apr 13;14(678):eabe4509. doi: 10.1126/scisignal.abe4509. Sci Signal. 2021. PMID: 33850054 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

