180-Nucleotide Duplication in the G Gene of Human metapneumovirus A2b Subgroup Strains Circulating in Yokohama City, Japan, since 2014
- PMID: 28352258
- PMCID: PMC5348506
- DOI: 10.3389/fmicb.2017.00402
180-Nucleotide Duplication in the G Gene of Human metapneumovirus A2b Subgroup Strains Circulating in Yokohama City, Japan, since 2014
Abstract
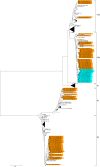
Human metapneumovirus (HMPV), a member of the family Paramyxoviridae, was first isolated in 2001. Seroepidemiological studies have shown that HMPV has been a major etiological agent of acute respiratory infections in humans for more than 50 years. Molecular epidemiological, genetic, and antigenetic evolutionary studies of HMPV will strengthen our understanding of the epidemic behavior of the virus and provide valuable insight for the control of HMPV and the development of vaccines and antiviral drugs against HMPV infection. In this study, the nucleotide sequence of and genetic variations in the G gene were analyzed in HMPV strains prevalent in Yokohama City, in the Kanto area, Japan, between January 2013 and June 2016. As a part of the National Epidemiological Surveillance of Infectious Diseases, Japan, 1308 clinical specimens (throat swabs, nasal swabs, nasal secretions, and nasal aspirate fluids) collected at 24 hospitals or clinics in Yokohama City were screened for 15 major respiratory viruses with a multiplex reverse transcription-PCR assay. HMPV was detected in 91 specimens, accounting for 7.0% of the total specimens, and the nucleotide sequences of the G genes of 84 HMPV strains were determined. Among these 84 strains, 6, 43, 10, and 25 strains were classified into subgroups A2a, A2b, B1, and B2, respectively. Approximately half the HMPV A2b subgroup strains detected since 2014 had a 180-nucleotide duplication (180nt-dup) in the G gene and clustered on a phylogenic tree with four classical 180nt-dup-lacking HMPV A2b strains prevalent between 2014 and 2015. The 180nt-dup causes a 60-amino-acid duplication (60aa-dup) in the G protein, creating 23-25 additional potential acceptor sites for O-linked sugars. Our data suggest that 180nt-dup occurred between 2011 and 2013 and that HMPV A2b strains with 180nt-dup (A2b180nt-dup HMPV) became major epidemic strains within 3 years. The detailed mechanism by which the A2b180nt-dup HMPV strains gained an advantage that allowed their efficient spread in the community and the effects of 60aa-dup on HMPV virulence must be clarified.
Keywords: G gene; Human metapneumovirus; duplication; molecular epidemiology; surveillance.
Figures




References
-
- Anon (2010). Pathogen surveillance system in Japan and Infectious Agents Surveillance Report (IASR). IASR 31 69–70.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

