Analysis of Evolutionary Processes of Species Jump in Waterfowl Parvovirus
- PMID: 28352261
- PMCID: PMC5349109
- DOI: 10.3389/fmicb.2017.00421
Analysis of Evolutionary Processes of Species Jump in Waterfowl Parvovirus
Abstract
Waterfowl parvoviruses are classified into goose parvovirus (GPV) and Muscovy duck parvovirus (MDPV) according to their antigenic features and host preferences. A novel duck parvovirus (NDPV), identified as a new variant of GPV, is currently infecting ducks, thus causing considerable economic loss. This study analyzed the molecular evolution and population dynamics of the emerging parvovirus capsid gene to investigate the evolutionary processes concerning the host shift of NDPV. Two important amino acids changes (Asn-489 and Asn-650) were identified in NDPV, which may be responsible for host shift of NDPV. Phylogenetic analysis indicated that the currently circulating NDPV originated from the GPV lineage. The Bayesian Markov chain Monte Carlo tree indicated that the NDPV diverged from GPV approximately 20 years ago. Evolutionary rate analyses demonstrated that GPV evolved with 7.674 × 10-4 substitutions/site/year, and the data for MDPV was 5.237 × 10-4 substitutions/site/year, whereas the substitution rate in NDPV branch was 2.25 × 10-3 substitutions/site/year. Meanwhile, viral population dynamics analysis revealed that the GPV major clade, including NDPV, grew exponentially at a rate of 1.717 year-1. Selection pressure analysis showed that most sites are subject to strong purifying selection and no positively selected sites were found in NDPV. The unique immune-epitopes in waterfowl parvovirus were also estimated, which may be helpful for the prediction of antibody binding sites against NDPV in ducks.
Keywords: Bayesian inference; epidemiology; evolution; phylogeny; species jump; waterfowl parvovirus.
Figures
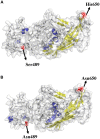

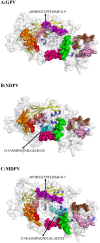
 represents NDPV strain and attenuated GPV strain.
represents NDPV strain and attenuated GPV strain.
Similar articles
-
Quantitative analysis of waterfowl parvoviruses in geese and Muscovy ducks by real-time polymerase chain reaction: correlation between age, clinical symptoms and DNA copy number of waterfowl parvoviruses.BMC Vet Res. 2012 Mar 15;8:29. doi: 10.1186/1746-6148-8-29. BMC Vet Res. 2012. PMID: 22420608 Free PMC article.
-
Molecular characterization of a novel Muscovy duck parvovirus isolate: evidence of recombination between classical MDPV and goose parvovirus strains.BMC Vet Res. 2017 Nov 9;13(1):327. doi: 10.1186/s12917-017-1238-6. BMC Vet Res. 2017. PMID: 29121936 Free PMC article.
-
Isolation and Genomic Characterization of a Duck-Origin GPV-Related Parvovirus from Cherry Valley Ducklings in China.PLoS One. 2015 Oct 14;10(10):e0140284. doi: 10.1371/journal.pone.0140284. eCollection 2015. PLoS One. 2015. PMID: 26465143 Free PMC article.
-
Identification of Goose-Origin Parvovirus as a Cause of Newly Emerging Beak Atrophy and Dwarfism Syndrome in Ducklings.J Clin Microbiol. 2016 Aug;54(8):1999-2007. doi: 10.1128/JCM.03244-15. Epub 2016 May 18. J Clin Microbiol. 2016. PMID: 27194692 Free PMC article.
-
Immunogenic Cross-Reactivity between Goose and Muscovy Duck Parvoviruses: Evaluation of Cross-Protection Provided by Mono- or Bivalent Vaccine.Vaccines (Basel). 2022 Aug 4;10(8):1255. doi: 10.3390/vaccines10081255. Vaccines (Basel). 2022. PMID: 36016142 Free PMC article.
Cited by
-
One-Step Multiplex Real-Time Fluorescent Quantitative Reverse Transcription PCR for Simultaneous Detection of Four Waterfowl Viruses.Microorganisms. 2024 Nov 25;12(12):2423. doi: 10.3390/microorganisms12122423. Microorganisms. 2024. PMID: 39770626 Free PMC article.
-
Advances in Research on Genetic Relationships of Waterfowl Parvoviruses.J Vet Res. 2021 Dec 2;65(4):391-399. doi: 10.2478/jvetres-2021-0063. eCollection 2021 Dec. J Vet Res. 2021. PMID: 35111991 Free PMC article.
-
Comparative pathogenicity of goose parvovirus across different epidemic lineages in ducklings and goslings.Virulence. 2025 Dec;16(1):2497904. doi: 10.1080/21505594.2025.2497904. Epub 2025 May 12. Virulence. 2025. PMID: 40302150 Free PMC article.
-
Attenuation of a Novel Goose Parvovirus Strain NMG21 via Serial Cell Passage.Viruses. 2025 Apr 25;17(5):618. doi: 10.3390/v17050618. Viruses. 2025. PMID: 40431630 Free PMC article.
-
Recombinant Muscovy Duck Parvovirus Led to Ileac Damage in Muscovy Ducklings.Viruses. 2022 Jul 3;14(7):1471. doi: 10.3390/v14071471. Viruses. 2022. PMID: 35891451 Free PMC article.
References
-
- Delaney N. F., Balenger S., Bonneaud C., Marx C. J., Hill G. E., Fergusonnoel N., et al. (2012). Correction: ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet. 8:e1002511 10.1371/journal.pgen.1002511 - DOI - PMC - PubMed
-
- Delano W. L. (2002). The PyMOL User’s Manual. Dpsm for Modeling Engineering Problems, Vol. 4 San Carlos, CA: DeLano Scientific, 148–149.
LinkOut - more resources
Full Text Sources
Other Literature Sources

