An Insight into microRNA156 Role in Salinity Stress Responses of Alfalfa
- PMID: 28352280
- PMCID: PMC5348497
- DOI: 10.3389/fpls.2017.00356
An Insight into microRNA156 Role in Salinity Stress Responses of Alfalfa
Abstract
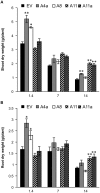
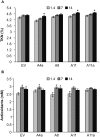
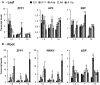
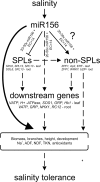
Salinity is one of the major abiotic stresses affecting alfalfa productivity. Developing salinity tolerant alfalfa genotypes could contribute to sustainable crop production. The functions of microRNA156 (miR156) have been investigated in several plant species, but so far, no studies have been published that explore the role of miR156 in alfalfa response to salinity stress. In this work, we studied the role of miR156 in modulating commercially important traits of alfalfa under salinity stress. Our results revealed that overexpression of miR156 increased biomass, number of branches and time to complete growth stages, while it reduced plant height under control and salinity stress conditions. We observed a miR156-related reduction in neutral detergent fiber under non-stress, and acid detergent fiber under mild salinity stress conditions. In addition, enhanced total Kjeldahl nitrogen content was recorded in miR156 overexpressing genotypes under severe salinity stress. Furthermore, alfalfa genotypes overexpressing miR156 exhibited an altered ion homeostasis under salinity conditions. Under severe salinity stress, miR156 downregulated SPL transcription factor family genes, modified expression of other important transcription factors, and downstream salt stress responsive genes. Taken together, our results reveal that miR156 plays a role in mediating physiological and transcriptional responses of alfalfa to salinity stress.
Keywords: alfalfa; biomass; ions; miR156; salt stress.
Figures




References
-
- Ahmed I. M., Dai H., Zheng W., Cao F., Zhang G., Sun D., et al. (2013). Genotypic differences in physiological characteristics in the tolerance to drought and salinity combined stress between Tibetan wild and cultivated barley. Plant. Physiol. Biochem. 63 49–60. 10.1016/j.plaphy.2012.11.004 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

